Nanoscale high-content analysis using compositional heterogeneities of single proteoliposomes
- PMID: 25086504
- PMCID: PMC4485457
- DOI: 10.1038/nmeth.3062
Nanoscale high-content analysis using compositional heterogeneities of single proteoliposomes
Abstract
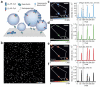
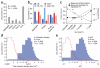
Proteoliposome reconstitution is a standard method to stabilize purified transmembrane proteins in membranes for structural and functional assays. Here we quantified intrareconstitution heterogeneities in single proteoliposomes using fluorescence microscopy. Our results suggest that compositional heterogeneities can severely skew ensemble-average proteoliposome measurements but also enable ultraminiaturized high-content screens. We took advantage of this screening capability to map the oligomerization energy of the β2-adrenergic receptor using ∼10(9)-fold less protein than conventional assays.
Figures

Similar articles
-
Single Proteoliposome High-Content Analysis Reveals Differences in the Homo-Oligomerization of GPCRs.Biophys J. 2018 Jul 17;115(2):300-312. doi: 10.1016/j.bpj.2018.05.036. Biophys J. 2018. PMID: 30021106 Free PMC article.
-
Single Vesicle Fluorescence-Bleaching Assay for Multi-Parameter Analysis of Proteoliposomes by Total Internal Reflection Fluorescence Microscopy.ACS Appl Mater Interfaces. 2022 Jul 6;14(26):29659-29667. doi: 10.1021/acsami.2c07454. Epub 2022 Jun 24. ACS Appl Mater Interfaces. 2022. PMID: 35748880 Free PMC article.
-
Spatial fluorescence cross-correlation spectroscopy.Appl Opt. 2006 Feb 20;45(6):1225-35. doi: 10.1364/ao.45.001225. Appl Opt. 2006. PMID: 16523786
-
Revealing G-protein-coupled receptor oligomerization at the single-molecule level through a nanoscopic lens: methods, dynamics and biological function.FEBS J. 2016 Apr;283(7):1197-217. doi: 10.1111/febs.13577. Epub 2015 Nov 28. FEBS J. 2016. PMID: 26509747 Review.
-
Single-molecule counting applied to the study of GPCR oligomerization.Biophys J. 2022 Sep 6;121(17):3175-3187. doi: 10.1016/j.bpj.2022.07.034. Epub 2022 Aug 3. Biophys J. 2022. PMID: 35927960 Free PMC article. Review.
Cited by
-
Micelle-Enhanced Bioorthogonal Labeling of Genetically Encoded Azido Groups on the Lipid-Embedded Surface of a GPCR.Chembiochem. 2015 Jun 15;16(9):1314-22. doi: 10.1002/cbic.201500030. Epub 2015 May 11. Chembiochem. 2015. PMID: 25962668 Free PMC article.
-
Direct visualization of single-molecule membrane protein interactions in living cells.PLoS Biol. 2018 Dec 13;16(12):e2006660. doi: 10.1371/journal.pbio.2006660. eCollection 2018 Dec. PLoS Biol. 2018. PMID: 30543635 Free PMC article.
-
In-Membrane Chemical Modification (IMCM) for Site-Specific Chromophore Labeling of GPCRs.Angew Chem Int Ed Engl. 2015 Dec 7;54(50):15246-9. doi: 10.1002/anie.201508506. Epub 2015 Nov 6. Angew Chem Int Ed Engl. 2015. PMID: 26545333 Free PMC article.
-
Permanent Photodynamic Cholecystokinin 1 Receptor Activation: Dimer-to-Monomer Conversion.Cell Mol Neurobiol. 2018 Aug;38(6):1283-1292. doi: 10.1007/s10571-018-0596-3. Epub 2018 Jun 4. Cell Mol Neurobiol. 2018. PMID: 29869099 Free PMC article.
-
The dimerization equilibrium of a ClC Cl(-)/H(+) antiporter in lipid bilayers.Elife. 2016 Aug 3;5:e17438. doi: 10.7554/eLife.17438. Elife. 2016. PMID: 27484630 Free PMC article.
References
-
- Rigaud JL, Lévy D. In: Methods in Enzymology: Liposomes, Part B. Duzgunes N, editor. Vol. 372. Elsevier; 2003. pp. 65–86. - PubMed
-
- Seddon AM, Curnow P, Booth PJ. Biochim. Biophys. Acta. 2004;1666:105–117. - PubMed
-
- Serebryany E, Zhu GA, Yan ECY. Biochim. Biophys. Acta. 2012;1818:225–233. - PubMed
-
- Niu SL, Doctrow B, Mitchell DC. Biochemistry. 2009;48:156–163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

