The growth-defense pivot: crisis management in plants mediated by LRR-RK surface receptors
- PMID: 25089011
- PMCID: PMC4177940
- DOI: 10.1016/j.tibs.2014.06.006
The growth-defense pivot: crisis management in plants mediated by LRR-RK surface receptors
Abstract
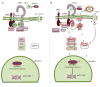
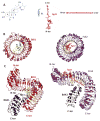
Plants must adapt to their environment and require mechanisms for sensing their surroundings and responding appropriately. An expanded family of more than 200 leucine-rich repeat (LRR) receptor kinases (LRR-RKs) transduces fluctuating and often contradictory signals from the environment into changes in nuclear gene expression. Two LRR-RKs, BRASSINOSTEROID INSENSITIVE 1 (BRI1), a steroid receptor, and FLAGELLIN SENSITIVE 2 (FLS2), an innate immune receptor that recognizes bacterial flagellin, act cooperatively to partition necessary growth-defense trade-offs. BRI1 and FLS2 share common signaling components and slightly different activation mechanisms. BRI1 and FLS2 are paradigms for understanding the signaling mechanisms of LRR-containing receptors in plants.
Keywords: LRR-RKs; cell surface signaling; crosstalk; innate immunity; plant growth; trade-offs.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

