A microRNA profile of human CD8(+) regulatory T cells and characterization of the effects of microRNAs on Treg cell-associated genes
- PMID: 25090912
- PMCID: PMC4440568
- DOI: 10.1186/s12967-014-0218-x
A microRNA profile of human CD8(+) regulatory T cells and characterization of the effects of microRNAs on Treg cell-associated genes
Abstract
Background: Recently, regulatory T (Treg) cells have gained interest in the fields of immunopathology, transplantation and oncoimmunology. Here, we investigated the microRNA expression profile of human natural CD8(+)CD25(+) Treg cells and the impact of microRNAs on molecules associated with immune regulation.
Methods: We purified human natural CD8(+) Treg cells and assessed the expression of FOXP3 and CTLA-4 by flow cytometry. We have also tested the ex vivo suppressive capacity of these cells in mixed leukocyte reactions. Using TaqMan low-density arrays and microRNA qPCR for validation, we could identify a microRNA 'signature' for CD8(+)CD25(+)FOXP3(+)CTLA-4(+) natural Treg cells. We used the 'TargetScan' and 'miRBase' bioinformatics programs to identify potential target sites for these microRNAs in the 3'-UTR of important Treg cell-associated genes.
Results: The human CD8(+)CD25(+) natural Treg cell microRNA signature includes 10 differentially expressed microRNAs. We demonstrated an impact of this signature on Treg cell biology by showing specific regulation of FOXP3, CTLA-4 and GARP gene expression by microRNA using site-directed mutagenesis and a dual-luciferase reporter assay. Furthermore, we used microRNA transduction experiments to demonstrate that these microRNAs impacted their target genes in human primary Treg cells ex vivo.
Conclusions: We are examining the biological relevance of this 'signature' by studying its impact on other important Treg cell-associated genes. These efforts could result in a better understanding of the regulation of Treg cell function and might reveal new targets for immunotherapy in immune disorders and cancer.
Figures

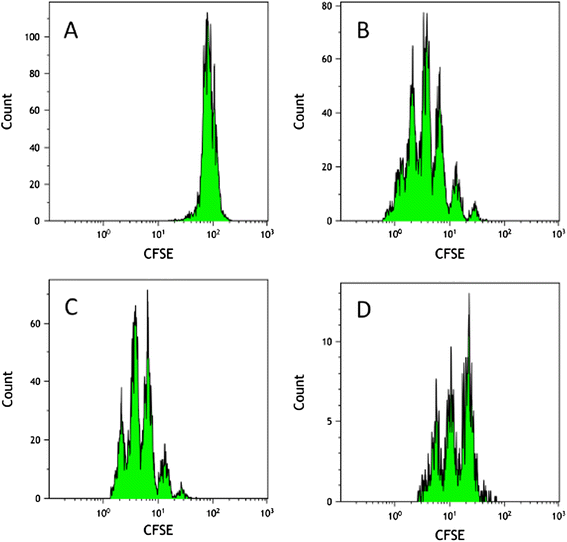

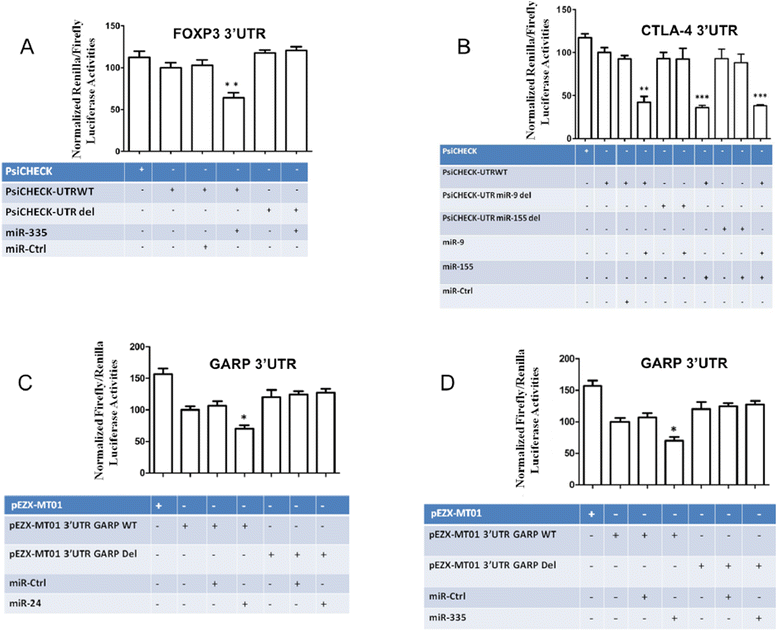
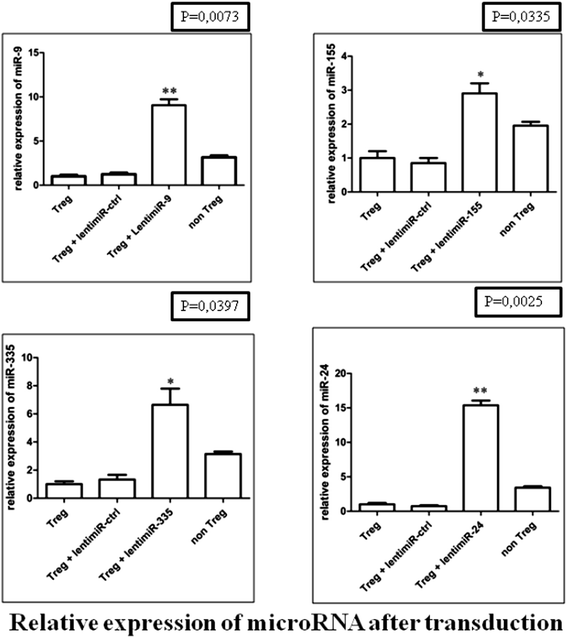
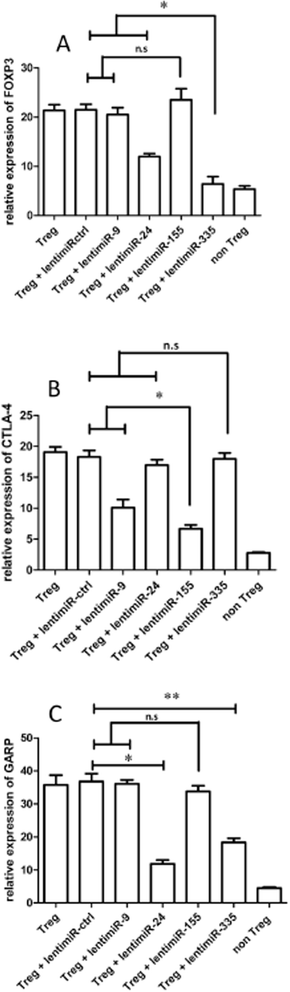
References
-
- Miller A, Lider O, Roberts AB, Sporn MB, Weiner HL. Suppressor T cells generated by oral tolerization to myelin basic protein suppress both in vitro and in vivo immune responses by the release of transforming growth factor beta after antigen-specific triggering. Proc Natl Acad Sci U S A. 1992;89(1):421–425. doi: 10.1073/pnas.89.1.421. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

