A joint analysis of metabolomics and genetics of breast cancer
- PMID: 25091696
- PMCID: PMC4187326
- DOI: 10.1186/s13058-014-0415-9
A joint analysis of metabolomics and genetics of breast cancer
Abstract
Introduction: Remodeling of cellular metabolism appears to be a consequence and possibly a cause of oncogenic transformation in human cancers. Specific aspects of altered tumor metabolism may be amenable to therapeutic intervention and could be coordinated with other targeted therapies. In breast cancer, the genetic landscape has been defined most comprehensively in efforts such as The Cancer Genome Atlas (TCGA). However, little is known about how alterations of tumor metabolism correlate with this landscape.
Methods: In total 25 cancers (23 fully analyzed by TCGA) and 5 normal breast specimens were analyzed by gas chromatography/mass spectrometry and liquid chromatography/mass spectrometry, quantitating 399 identifiable metabolites.
Results: We found strong differences correlated with hormone receptor status with 18% of the metabolites elevated in estrogen receptor negative (ER-) cancers compared to estrogen receptor positive (ER+) including many glycolytic and glycogenolytic intermediates consistent with increased Warburg effects. Glutathione (GSH) pathway components were also elevated in ER- tumors consistent with an increased requirement for handling higher levels of oxidative stress. Additionally, ER- tumors had high levels of the oncometabolite 2-hydroxyglutarate (2-HG) and the immunomodulatory tryptophan metabolite kynurenine. Kynurenine levels were correlated with the expression of tryptophan-degrading enzyme (IDO1). However, high levels of 2-HG were not associated with somatic mutations or expression levels of IDH1 or IDH2. BRCA1 mRNA levels were positively associated with coenzyme A, acetyl coenzyme A, and GSH and negatively associated with multiple lipid species, supporting the regulation of ACC1 and NRF2 by BRCA1. Different driver mutations were associated with distinct patterns of specific metabolites, such as lower levels of several lipid-glycerophosphocholines in tumors with mutated TP53. A strong metabolomic signature associated with proliferation rate was also observed; the metabolites in this signature overlap broadly with metabolites that define ER status as receptor status and proliferation rate were correlated.
Conclusions: The addition of metabolomic profiles to the public domain TCGA dataset provides an important new tool for discovery and hypothesis testing of the genetic regulation of tumor metabolism. Particular sets of metabolites may reveal insights into the metabolic dysregulation that underlie the heterogeneity of breast cancer.
Figures



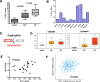
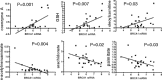

References
-
- Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, Mates JM, Alonso FJ, Wang C, Seo Y, Chen X, Bishop JM. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

