Genomic responses in mouse models greatly mimic human inflammatory diseases
- PMID: 25092317
- PMCID: PMC4313832
- DOI: 10.1073/pnas.1401965111
Genomic responses in mouse models greatly mimic human inflammatory diseases
Erratum in
-
Correction for Takao and Miyakawa, Genomic responses in mouse models greatly mimic human inflammatory diseases.Proc Natl Acad Sci U S A. 2015 Mar 10;112(10):E1163-7. doi: 10.1073/pnas.1502188112. Epub 2015 Feb 17. Proc Natl Acad Sci U S A. 2015. PMID: 25691751 Free PMC article. No abstract available.
Abstract
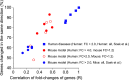
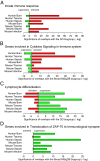
The use of mice as animal models has long been considered essential in modern biomedical research, but the role of mouse models in research was challenged by a recent report that genomic responses in mouse models poorly mimic human inflammatory diseases. Here we reevaluated the same gene expression datasets used in the previous study by focusing on genes whose expression levels were significantly changed in both humans and mice. Contrary to the previous findings, the gene expression levels in the mouse models showed extraordinarily significant correlations with those of the human conditions (Spearman's rank correlation coefficient: 0.43-0.68; genes changed in the same direction: 77-93%; P = 6.5 × 10(-11) to 1.2 × 10(-35)). Moreover, meta-analysis of those datasets revealed a number of pathways/biogroups commonly regulated by multiple conditions in humans and mice. These findings demonstrate that gene expression patterns in mouse models closely recapitulate those in human inflammatory conditions and strongly argue for the utility of mice as animal models of human disorders.
Keywords: burn; inflammation; sepsis; transcriptome analysis; trauma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Reply to Warren et al. and Shay et al.: Commonalities across species do exist and are potentially important.Proc Natl Acad Sci U S A. 2015 Jan 27;112(4):E347-8. doi: 10.1073/pnas.1417369111. Epub 2014 Dec 24. Proc Natl Acad Sci U S A. 2015. PMID: 25540421 Free PMC article. No abstract available.
-
Mice are not men.Proc Natl Acad Sci U S A. 2015 Jan 27;112(4):E345. doi: 10.1073/pnas.1414857111. Epub 2014 Dec 24. Proc Natl Acad Sci U S A. 2015. PMID: 25540422 Free PMC article. No abstract available.
-
Genomic responses to inflammation in mouse models mimic humans: we concur, apples to oranges comparisons won't do.Proc Natl Acad Sci U S A. 2015 Jan 27;112(4):E346. doi: 10.1073/pnas.1416629111. Epub 2014 Dec 24. Proc Natl Acad Sci U S A. 2015. PMID: 25540423 Free PMC article. No abstract available.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

