Convex lens-induced nanoscale templating
- PMID: 25092333
- PMCID: PMC4169971
- DOI: 10.1073/pnas.1321089111
Convex lens-induced nanoscale templating
Abstract
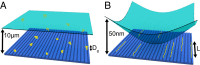
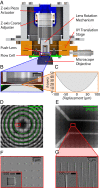
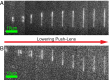
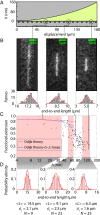
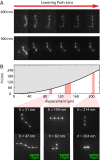
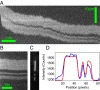
We demonstrate a new platform, convex lens-induced nanoscale templating (CLINT), for dynamic manipulation and trapping of single DNA molecules. In the CLINT technique, the curved surface of a convex lens is used to deform a flexible coverslip above a substrate containing embedded nanotopography, creating a nanoscale gap that can be adjusted during an experiment to confine molecules within the embedded nanostructures. Critically, CLINT has the capability of transforming a macroscale flow cell into a nanofluidic device without the need for permanent direct bonding, thus simplifying sample loading, providing greater accessibility of the surface for functionalization, and enabling dynamic manipulation of confinement during device operation. Moreover, as DNA molecules present in the gap are driven into the embedded topography from above, CLINT eliminates the need for the high pressures or electric fields required to load DNA into direct-bonded nanofluidic devices. To demonstrate the versatility of CLINT, we confine DNA to nanogroove and nanopit structures, demonstrating DNA nanochannel-based stretching, denaturation mapping, and partitioning/trapping of single molecules in multiple embedded cavities. In particular, using ionic strengths that are in line with typical biological buffers, we have successfully extended DNA in sub-30-nm nanochannels, achieving high stretching (90%) that is in good agreement with Odijk deflection theory, and we have mapped genomic features using denaturation analysis.
Keywords: CLIC imaging; genomic mapping; nanotechnology; polymer confinement; single-molecule manipulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Comment in
-
How to get into that "room at the bottom".Proc Natl Acad Sci U S A. 2014 Sep 16;111(37):13249-50. doi: 10.1073/pnas.1414387111. Epub 2014 Sep 3. Proc Natl Acad Sci U S A. 2014. PMID: 25187558 Free PMC article. No abstract available.
References
-
- Reisner W, Pedersen JN, Austin RH. DNA confinement in nanochannels: Physics and biological applications. Rep Prog Phys. 2012;75(10):106601–106610. - PubMed
-
- Bruus H, Microfuidics C. 2008. Theoretical Microfluidics (Oxford University Press, Oxford)
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

