Nfil3 is required for the development of all innate lymphoid cell subsets
- PMID: 25092873
- PMCID: PMC4144736
- DOI: 10.1084/jem.20140145
Nfil3 is required for the development of all innate lymphoid cell subsets
Abstract
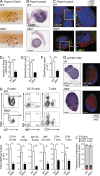
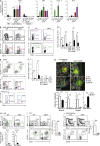
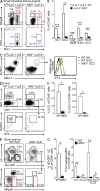
Innate lymphoid cell (ILC) populations protect against infection and are essential for lymphoid tissue formation and tissue remodeling after damage. Nfil3 is implicated in the function of adaptive immune lineages and NK cell development, but it is not yet known if Nfil3 regulates other innate lymphoid lineages. Here, we identify that Nfil3 is essential for the development of Peyer's patches and ILC2 and ILC3 subsets. Loss of Nfil3 selectively reduced Peyer's patch formation and was accompanied by impaired recruitment and distribution of lymphocytes within the patches. ILC subsets exhibited high Nfil3 expression and genetic deletion of Nfil3 severely compromised the development of all subsets. Subsequently, Nfil3(-/-) mice were highly susceptible to disease when challenged with inflammatory or infectious agents. Thus, we demonstrate that Nfil3 is a key regulator of the development of ILC subsets essential for immune protection in the lung and gut.
© 2014 Seillet et al.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

