A functional genomic screen for evolutionarily conserved genes required for lifespan and immunity in germline-deficient C. elegans
- PMID: 25093668
- PMCID: PMC4122342
- DOI: 10.1371/journal.pone.0101970
A functional genomic screen for evolutionarily conserved genes required for lifespan and immunity in germline-deficient C. elegans
Abstract
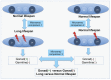
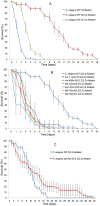
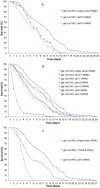
The reproductive system regulates lifespan in insects, nematodes and vertebrates. In Caenorhabditis elegans removal of germline increases lifespan by 60% which is dependent upon insulin signaling, nuclear hormone signaling, autophagy and fat metabolism and their microRNA-regulators. Germline-deficient C. elegans are also more resistant to various bacterial pathogens but the underlying molecular mechanisms are largely unknown. Firstly, we demonstrate that previously identified genes that regulate the extended lifespan of germline-deficient C. elegans (daf-2, daf-16, daf-12, tcer-1, mir-7.1 and nhr-80) are also essential for resistance to the pathogenic bacterium Xenorhabdus nematophila. We then use a novel unbiased approach combining laser cell ablation, whole genome microarrays, RNAi screening and exposure to X. nematophila to generate a comprehensive genome-wide catalog of genes potentially required for increased lifespan and innate immunity in germline-deficient C. elegans. We find 3,440 genes to be upregulated in C. elegans germline-deficient animals in a gonad dependent manner, which are significantly enriched for genes involved in insulin signaling, fatty acid desaturation, translation elongation and proteasome complex function. Using RNAi against a subset of 150 candidate genes selected from the microarray results, we show that the upregulated genes such as transcription factor DAF-16/FOXO, the PTEN homolog lipid phosphatase DAF-18 and several components of the proteasome complex (rpn-6.1, rpn-7, rpn-9, rpn-10, rpt-6, pbs-3 and pbs-6) are essential for both lifespan and immunity of germline deficient animals. We also identify a novel role for genes including par-5 and T12G3.6 in both lifespan-extension and increased survival on X. nematophila. From an evolutionary perspective, most of the genes differentially expressed in germline deficient C. elegans also show a conserved expression pattern in germline deficient Pristionchus pacificus, a nematode species that diverged from C. elegans 250-400 MYA.
Conflict of interest statement
Figures


Similar articles
-
Genome-wide analysis of germline signaling genes regulating longevity and innate immunity in the nematode Pristionchus pacificus.PLoS Pathog. 2012;8(8):e1002864. doi: 10.1371/journal.ppat.1002864. Epub 2012 Aug 9. PLoS Pathog. 2012. PMID: 22912581 Free PMC article.
-
System wide analysis of the evolution of innate immunity in the nematode model species Caenorhabditis elegans and Pristionchus pacificus.PLoS One. 2012;7(9):e44255. doi: 10.1371/journal.pone.0044255. Epub 2012 Sep 28. PLoS One. 2012. PMID: 23028509 Free PMC article.
-
The somatic reproductive tissues of C. elegans promote longevity through steroid hormone signaling.PLoS Biol. 2010 Aug 31;8(8):e1000468. doi: 10.1371/journal.pbio.1000468. PLoS Biol. 2010. PMID: 20824162 Free PMC article.
-
A pathway that links reproductive status to lifespan in Caenorhabditis elegans.Ann N Y Acad Sci. 2010 Aug;1204:156-62. doi: 10.1111/j.1749-6632.2010.05640.x. Ann N Y Acad Sci. 2010. PMID: 20738286 Review.
-
DAF-16: FOXO in the Context of C. elegans.Curr Top Dev Biol. 2018;127:1-21. doi: 10.1016/bs.ctdb.2017.11.007. Epub 2018 Feb 2. Curr Top Dev Biol. 2018. PMID: 29433733 Review.
Cited by
-
ELLI-1, a novel germline protein, modulates RNAi activity and P-granule accumulation in Caenorhabditis elegans.PLoS Genet. 2017 Feb 9;13(2):e1006611. doi: 10.1371/journal.pgen.1006611. eCollection 2017 Feb. PLoS Genet. 2017. PMID: 28182654 Free PMC article.
-
Genetic Screen Reveals Link between the Maternal Effect Sterile Gene mes-1 and Pseudomonas aeruginosa-induced Neurodegeneration in Caenorhabditis elegans.J Biol Chem. 2015 Dec 4;290(49):29231-9. doi: 10.1074/jbc.M115.674259. Epub 2015 Oct 16. J Biol Chem. 2015. PMID: 26475858 Free PMC article.
-
Forward genetic screening identifies novel roles for N-terminal acetyltransferase C and histone deacetylase in C. elegans development.Sci Rep. 2022 Sep 30;12(1):16438. doi: 10.1038/s41598-022-20361-x. Sci Rep. 2022. PMID: 36180459 Free PMC article.
-
Transcriptomic evidence for a trade-off between germline proliferation and immunity in Drosophila.Evol Lett. 2021 Oct 21;5(6):644-656. doi: 10.1002/evl3.261. eCollection 2021 Dec. Evol Lett. 2021. PMID: 34917403 Free PMC article.
-
The Energy Maintenance Theory of Aging: Maintaining Energy Metabolism to Allow Longevity.Bioessays. 2018 Aug;40(8):e1800005. doi: 10.1002/bies.201800005. Epub 2018 Jun 14. Bioessays. 2018. PMID: 29901833 Free PMC article. Review.
References
-
- Smith JM (1958) The Effects of Temperature and of Egg-Laying on the Longevity of Drosophila Subobscura. J Exp Biol 35: 832–842.
-
- Robertson OH (1961) Relation of gonadal maturation to length of life in Pacific salmon. Fed. Proc. (Suppl 8): 29–30. - PubMed
-
- Hamilton JB (1965) Relationship of castration, spaying, and sex to survival and duration of life in domestic cats. J Gerontol 20: 96–104. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

