Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1
- PMID: 25096180
- PMCID: PMC4166716
- DOI: 10.1152/ajpendo.00204.2014
Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1
Abstract
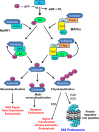
Muscle RING finger 1 (MuRF1) and muscle atrophy F-box (MAFbx)/atrogin-1 were identified more than 10 years ago as two muscle-specific E3 ubiquitin ligases that are increased transcriptionally in skeletal muscle under atrophy-inducing conditions, making them excellent markers of muscle atrophy. In the past 10 years much has been published about MuRF1 and MAFbx with respect to their mRNA expression patterns under atrophy-inducing conditions, their transcriptional regulation, and their putative substrates. However, much remains to be learned about the physiological role of both genes in the regulation of mass and other cellular functions in striated muscle. Although both MuRF1 and MAFbx are enriched in skeletal, cardiac, and smooth muscle, this review will focus on the current understanding of MuRF1 and MAFbx in skeletal muscle, highlighting the critical questions that remain to be answered.
Keywords: atrogenes; muscle RING finger 1; muscle atrophy F-box; muscle sparing; protein quality control; ubiquitin proteasome system.
Figures

References
-
- Adams V, Mangner N, Gasch A, Krohne C, Gielen S, Hirner S, Thierse HJ, Witt CC, Linke A, Schuler G, Labeit S. Induction of MuRF1 is essential for TNF-alpha-induced loss of muscle function in mice. J Mol Biol 384: 48–59, 2008 - PubMed
-
- Allen DL, Cleary AS, Lindsay SF, Loh AS, Reed JM. Myostatin expression is increased by food deprivation in a muscle-specific manner and contributes to muscle atrophy during prolonged food deprivation in mice. J Appl Physiol 109: 692–701, 2010 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

