Comparative genomics of 274 Vibrio cholerae genomes reveals mobile functions structuring three niche dimensions
- PMID: 25096633
- PMCID: PMC4141962
- DOI: 10.1186/1471-2164-15-654
Comparative genomics of 274 Vibrio cholerae genomes reveals mobile functions structuring three niche dimensions
Abstract
Background: Vibrio cholerae is a globally dispersed pathogen that has evolved with humans for centuries, but also includes non-pathogenic environmental strains. Here, we identify the genomic variability underlying this remarkable persistence across the three major niche dimensions space, time, and habitat.
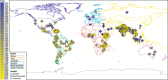
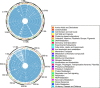
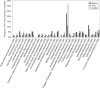
Results: Taking an innovative approach of genome-wide association applicable to microbial genomes (GWAS-M), we classify 274 complete V. cholerae genomes by niche, including 39 newly sequenced for this study with the Ion Torrent DNA-sequencing platform. Niche metadata were collected for each strain and analyzed together with comprehensive annotations of genetic and genomic attributes, including point mutations (single-nucleotide polymorphisms, SNPs), protein families, functions and prophages.
Conclusions: Our analysis revealed that genomic variations, in particular mobile functions including phages, prophages, transposable elements, and plasmids underlie the metadata structuring in each of the three niche dimensions. This underscores the role of phages and mobile elements as the most rapidly evolving elements in bacterial genomes, creating local endemicity (space), leading to temporal divergence (time), and allowing the invasion of new habitats. Together, we take a data-driven approach for comparative functional genomics that exploits high-volume genome sequencing and annotation, in conjunction with novel statistical and machine learning analyses to identify connections between genotype and phenotype on a genome-wide scale.
Figures

References
-
- Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, Croucher NJ, Choi SY, Harris SR, Lebens M, Niyogi SK, Kim EJ, Ramamurthy T, Chun J, Wood JL, Clemens JD, Czerkinsky C, Nair GB, Holmgren J, Parkhill J, Dougan G. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477(7365):462–465. doi: 10.1038/nature10392. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

