Coronaviruses induce entry-independent, continuous macropinocytosis
- PMID: 25096879
- PMCID: PMC4128357
- DOI: 10.1128/mBio.01340-14
Coronaviruses induce entry-independent, continuous macropinocytosis
Abstract
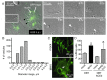
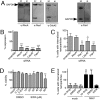
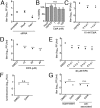
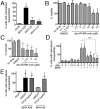
Macropinocytosis is exploited by many pathogens for entry into cells. Coronaviruses (CoVs) such as severe acute respiratory syndrome (SARS) CoV and Middle East respiratory syndrome CoV are important human pathogens; however, macropinocytosis during CoV infection has not been investigated. We demonstrate that the CoVs SARS CoV and murine hepatitis virus (MHV) induce macropinocytosis, which occurs late during infection, is continuous, and is not associated with virus entry. MHV-induced macropinocytosis results in vesicle internalization, as well as extended filopodia capable of fusing with distant cells. MHV-induced macropinocytosis requires fusogenic spike protein on the cell surface and is dependent on epidermal growth factor receptor activation. Inhibition of macropinocytosis reduces supernatant viral titers and syncytia but not intracellular virus titers. These results indicate that macropinocytosis likely facilitates CoV infection through enhanced cell-to-cell spreading. Our studies are the first to demonstrate virus use of macropinocytosis for a role other than entry and suggest a much broader potential exploitation of macropinocytosis in virus replication and host interactions. Importance: Coronaviruses (CoVs), including severe acute respiratory syndrome (SARS) CoV and Middle East respiratory syndrome CoV, are critical emerging human pathogens. Macropinocytosis is induced by many pathogens to enter host cells, but other functions for macropinocytosis in virus replication are unknown. In this work, we show that CoVs induce a macropinocytosis late in infection that is continuous, independent from cell entry, and associated with increased virus titers and cell fusion. Murine hepatitis virus macropinocytosis requires a fusogenic virus spike protein and signals through the epidermal growth factor receptor and the classical macropinocytosis pathway. These studies demonstrate CoV induction of macropinocytosis for a purpose other than entry and indicate that viruses likely exploit macropinocytosis at multiple steps in replication and pathogenesis.
Copyright © 2014 Freeman et al.
Figures



References
-
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, SARS Working Group 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953–1966. 10.1056/NEJMoa030781 - DOI - PubMed
-
- Kuiken T, Fouchier RA, Schutten M, Rimmelzwaan GF, van Amerongen G, van Riel D, Laman JD, de Jong T, van Doornum G, Lim W, Ling AE, Chan PK, Tam JS, Zambon MC, Gopal R, Drosten C, van der Werf S, Escriou N, Manuguerra JC, Stöhr K, Peiris JS, Osterhaus AD. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362:263–270. 10.1016/S0140-6736(03)13967-0 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
