Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis
- PMID: 25097262
- PMCID: PMC4143034
- DOI: 10.1073/pnas.1404629111
Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis
Abstract
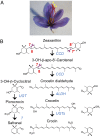
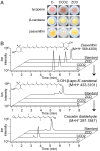
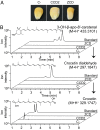
Crocus sativus stigmas are the source of the saffron spice and accumulate the apocarotenoids crocetin, crocins, picrocrocin, and safranal, responsible for its color, taste, and aroma. Through deep transcriptome sequencing, we identified a novel dioxygenase, carotenoid cleavage dioxygenase 2 (CCD2), expressed early during stigma development and closely related to, but distinct from, the CCD1 dioxygenase family. CCD2 is the only identified member of a novel CCD clade, presents the structural features of a bona fide CCD, and is able to cleave zeaxanthin, the presumed precursor of saffron apocarotenoids, both in Escherichia coli and in maize endosperm. The cleavage products, identified through high-resolution mass spectrometry and comigration with authentic standards, are crocetin dialdehyde and crocetin, respectively. In vitro assays show that CCD2 cleaves sequentially the 7,8 and 7',8' double bonds adjacent to a 3-OH-β-ionone ring and that the conversion of zeaxanthin to crocetin dialdehyde proceeds via the C30 intermediate 3-OH-β-apo-8'-carotenal. In contrast, zeaxanthin cleavage dioxygenase (ZCD), an enzyme previously claimed to mediate crocetin formation, did not cleave zeaxanthin or 3-OH-β-apo-8'-carotenal in the test systems used. Sequence comparison and structure prediction suggest that ZCD is an N-truncated CCD4 form, lacking one blade of the β-propeller structure conserved in all CCDs. These results constitute strong evidence that CCD2 catalyzes the first dedicated step in crocin biosynthesis. Similar to CCD1, CCD2 has a cytoplasmic localization, suggesting that it may cleave carotenoids localized in the chromoplast outer envelope.
Keywords: symmetric carotenoid cleavage; β-citraurin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Fernandez JA, Pandalai SG. Biology, biotechnology and biomedicine of saffron. Recent Res Dev Plant Sci. 2004;2:127–159.
-
- Caballero-Ortega H, Pereda-Miranda R, Abdullaev FI. HPLC quantification of major active components from 11 different saffron (Crocus sativus L.) sources. Food Chem. 2007;100(3):1126–1131.
-
- Pfander H, Schurtenberger H. Biosynthesis of C20-carotenoids in Crocus sativus. Phytochemistry. 1982;21(5):1039–1042.
-
- Moraga AR, Nohales PF, Pérez JA, Gómez-Gómez L. Glucosylation of the saffron apocarotenoid crocetin by a glucosyltransferase isolated from Crocus sativus stigmas. Planta. 2004;219(6):955–966. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

