Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential
- PMID: 25097266
- PMCID: PMC4151758
- DOI: 10.1073/pnas.1413299111
Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential
Abstract
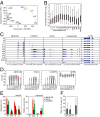
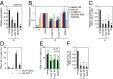
Pluripotency can be induced in somatic cells by overexpressing transcription factors, including POU class 5 homeobox 1 (OCT3/4), sex determining region Y-box 2 (SOX2), Krüppel-like factor 4 (KLF4), and myelocytomatosis oncogene (c-MYC). However, some induced pluripotent stem cells (iPSCs) exhibit defective differentiation and inappropriate maintenance of pluripotency features. Here we show that dynamic regulation of human endogenous retroviruses (HERVs) is important in the reprogramming process toward iPSCs, and in re-establishment of differentiation potential. During reprogramming, OCT3/4, SOX2, and KLF4 transiently hyperactivated LTR7s--the long-terminal repeats of HERV type-H (HERV-H)--to levels much higher than in embryonic stem cells by direct occupation of LTR7 sites genome-wide. Knocking down LTR7s or long intergenic non-protein coding RNA, regulator of reprogramming (lincRNA-RoR), a HERV-H-driven long noncoding RNA, early in reprogramming markedly reduced the efficiency of iPSC generation. KLF4 and LTR7 expression decreased to levels comparable with embryonic stem cells once reprogramming was complete, but failure to resuppress KLF4 and LTR7s resulted in defective differentiation. We also observed defective differentiation and LTR7 activation when iPSCs had forced expression of KLF4. However, when aberrantly expressed KLF4 or LTR7s were suppressed in defective iPSCs, normal differentiation was restored. Thus, a major mechanism by which OCT3/4, SOX2, and KLF4 promote human iPSC generation and reestablish potential for differentiation is by dynamically regulating HERV-H LTR7s.
Keywords: epigenetics; evolution; retrotransposon.
Conflict of interest statement
Conflict of interest statement: S.Y. is a member without salary of the scientific advisory board of iPS Academia Japan.
Figures




References
-
- Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. - PubMed
-
- Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. - PubMed
-
- Lander ES, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

