TLR9 ligands induce S100A8 in macrophages via a STAT3-dependent pathway which requires IL-10 and PGE2
- PMID: 25098409
- PMCID: PMC4123874
- DOI: 10.1371/journal.pone.0103629
TLR9 ligands induce S100A8 in macrophages via a STAT3-dependent pathway which requires IL-10 and PGE2
Abstract
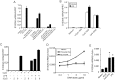
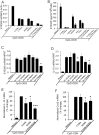
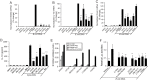
S100A8 and S100A9 are highly-expressed calcium-binding proteins in neutrophils and monocytes, and in subsets of macrophages in inflammatory lesions. Unmethylated CpG motifs found in bacterial and viral DNA are potent activators of innate immunity via Toll-like receptor 9 (TLR9). S100A8, but not S100A9, mRNA and protein was directly induced by CpG-DNA in murine and human macrophages. Induction in murine macrophages peaked at 16 h. CpG-DNA-induced S100A8 required de novo protein synthesis; IL-10 and Prostaglandin E2 (PGE2) synergistically enhanced expression and promoted earlier gene induction. Inhibitors of endogenous IL-10, PGE2, and the E prostanoid (EP) 4 receptor strongly suppressed S100A8 expression, particularly when combined. Thus, S100A8 induction by E. coli DNA required both IL-10 and PGE2/EP4 signaling. The MAPKs, PI3K and JAK pathways were essential, whereas ERK1/2 appeared to play a direct role. S100A8 induction by CpG-DNA was controlled at the transcriptional level. The promoter region responsible for activation, either directly, or indirectly via IL-10 and PGE2, was located within a -178 to -34-bp region and required STAT3 binding. Because of the robust links connecting IL-10 and PGE2 with an anti-inflammatory macrophage phenotype, the induction profile of S100A8 strongly indicates a role for this protein in resolution of inflammation.
Conflict of interest statement
Figures



References
-
- Takeuchi O, Akira S (2010) Pattern recognition receptors and inflammation. Cell 140: 805–820. - PubMed
-
- Medzhitov R, Horng T (2009) Transcriptional control of the inflammatory response. Nat Rev Immunol 9: 692–703. - PubMed
-
- Krieg AM (2002) CpG motifs in bacterial DNA and their immune effects. Annu Rev Immunol 20: 709–760. - PubMed
-
- Haas T, Metzger J, Schmitz F, Heit A, Muller T, et al. (2008) The DNA sugar backbone 2′ deoxyribose determines toll-like receptor 9 activation. Immunity 28: 315–323. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

