Integration of binding peptide selection and multifunctional particles as tool-box for capture of soluble proteins in serum
- PMID: 25100324
- PMCID: PMC4233757
- DOI: 10.1098/rsif.2014.0718
Integration of binding peptide selection and multifunctional particles as tool-box for capture of soluble proteins in serum
Abstract
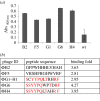
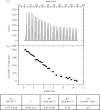
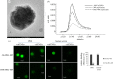
In this paper, we report on a general approach for the detection of a specific tumoural biomarker directly in serum. Such detection is made possible using a protein-binding peptide selected through an improved phage display technique and then conjugated to engineered microparticles (MPs). Protein biomarkers represent an unlimited source of information for non-invasive diagnostic and prognostic tests; MP-based assays are becoming largely used in manipulation of soluble biomarkers, but their direct use in serum is hampered by the complex biomolecular environment. Our technique overcomes the current limitations as it produces a selective MP--engineered with an antifouling layer--that 'captures' the relevant protein staying impervious to the background. Our system succeeds in fishing-out the human tumour necrosis factor alpha directly in serum with a high selectivity degree. Our method could have great impact in soluble protein manipulation and detection for a wide variety of diagnostic applications.
Keywords: microparticles for diagnosis; phage display selection; soluble cancer biomarker.
© 2014 The Author(s) Published by the Royal Society. All rights reserved.
Figures



Similar articles
-
Modification of phage display technique for improved screening of high-affinity binding peptides.J Biotechnol. 2019 Jan 10;289:88-92. doi: 10.1016/j.jbiotec.2018.11.020. Epub 2018 Nov 26. J Biotechnol. 2019. PMID: 30496775
-
Identification of Soft Matter Binding Peptide Ligands Using Phage Display.Bioconjug Chem. 2015 Oct 21;26(10):2002-15. doi: 10.1021/acs.bioconjchem.5b00377. Epub 2015 Sep 8. Bioconjug Chem. 2015. PMID: 26275106 Review.
-
A combination of in vitro techniques for efficient discovery of functional monoclonal antibodies against human CXC chemokine receptor-2 (CXCR2).MAbs. 2014;6(6):1415-24. doi: 10.4161/mabs.36237. MAbs. 2014. PMID: 25484047 Free PMC article.
-
Phage display selection of tight specific binding variants from a hyperthermostable Sso7d scaffold protein library.FEBS J. 2016 Apr;283(7):1351-67. doi: 10.1111/febs.13674. Epub 2016 Mar 6. FEBS J. 2016. PMID: 26835881
-
Advance in phage display technology for bioanalysis.Biotechnol J. 2016 Jun;11(6):732-45. doi: 10.1002/biot.201500458. Epub 2016 Apr 8. Biotechnol J. 2016. PMID: 27061133 Review.
Cited by
-
Bioengineering Microgels and Hydrogel Microparticles for Sensing Biomolecular Targets.Gels. 2017 May 30;3(2):20. doi: 10.3390/gels3020020. Gels. 2017. PMID: 30920517 Free PMC article. Review.
-
Supramolecular Hydrogelators and Hydrogels: From Soft Matter to Molecular Biomaterials.Chem Rev. 2015 Dec 23;115(24):13165-307. doi: 10.1021/acs.chemrev.5b00299. Epub 2015 Dec 8. Chem Rev. 2015. PMID: 26646318 Free PMC article. Review.
-
Current status, challenges and prospects of antifouling materials for oncology applications.Front Oncol. 2024 May 8;14:1391293. doi: 10.3389/fonc.2024.1391293. eCollection 2024. Front Oncol. 2024. PMID: 38779096 Free PMC article. Review.
-
Easy Surface Functionalization and Bioconjugation of Peptides as Capture Agents of a Microfluidic Biosensing Platform for Multiplex Assay in Serum.Bioconjug Chem. 2021 Aug 18;32(8):1593-1601. doi: 10.1021/acs.bioconjchem.1c00146. Epub 2021 Jun 11. Bioconjug Chem. 2021. PMID: 34114801 Free PMC article.
References
-
- Barh D, Agte V, Dhawan D, Agte V, Padh H. 2012. Cancer biomarkers for diagnosis, prognosis and therapy. In Molecular and cellular therapeutics (eds D Whitehouse, R Rapley), pp. 18–68. Chichester, UK: John Wiley & Sons, Ltd.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

