Activation of neuronal NMDA receptors triggers transient ATP-mediated microglial process outgrowth
- PMID: 25100586
- PMCID: PMC6802598
- DOI: 10.1523/JNEUROSCI.0405-14.2014
Activation of neuronal NMDA receptors triggers transient ATP-mediated microglial process outgrowth
Abstract
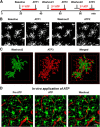
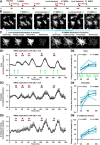
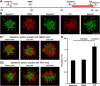
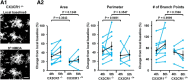
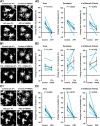
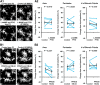
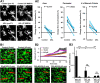
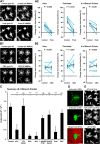
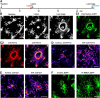
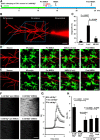
Microglia are morphologically dynamic cells that rapidly extend their processes in response to various stimuli including extracellular ATP. In this study, we tested the hypothesis that stimulation of neuronal NMDARs trigger ATP release leading to communication with microglia. We used acute mouse hippocampal brain slices and two-photon laser scanning microscopy to study microglial dynamics and developed a novel protocol for fixation and immunolabeling of microglia processes. Similar to direct topical ATP application in vivo, short multiple applications of NMDA triggered transient microglia process outgrowth that was reversible and repeatable indicating that this was not due to excitotoxic damage. Stimulation of NMDAR was required as NMDAR antagonists, but not blockers of AMPA/kainate receptors or voltage-gated sodium channels, prevented microglial outgrowth. We report that ATP release, secondary to NMDAR activation, was the key mediator of this neuron-microglia communication as both blocking purinergic receptors and inhibiting hydrolysis of ATP to prevent locally generated gradients abolished outgrowth. Pharmacological and genetic analyses showed that the NMDA-triggered microglia process extension was independent of Pannexin 1, the ATP releasing channels, ATP release from astrocytes via connexins, and nitric oxide generation. Finally, using whole-cell patch clamping we demonstrate that activation of dendritic NMDAR on single neurons is sufficient to trigger microglia process outgrowth. Our results suggest that dendritic neuronal NMDAR activation triggers ATP release via a Pannexin 1-independent manner that induces outgrowth of microglia processes. This represents a novel uncharacterized form of neuron-microglial communication mediated by ATP.
Keywords: ATP purinergic receptors; NMDA glutamate receptors; Pannexin; microglia process motility; synaptic surveillance.
Copyright © 2014 the authors 0270-6474/14/3410511-17$15.00/0.
Figures

Similar articles
-
Neuronal hyperactivity recruits microglial processes via neuronal NMDA receptors and microglial P2Y12 receptors after status epilepticus.J Neurosci. 2014 Aug 6;34(32):10528-40. doi: 10.1523/JNEUROSCI.0416-14.2014. J Neurosci. 2014. PMID: 25100587 Free PMC article.
-
Purinergic signaling induces cyclooxygenase-1-dependent prostanoid synthesis in microglia: roles in the outcome of excitotoxic brain injury.PLoS One. 2011;6(10):e25916. doi: 10.1371/journal.pone.0025916. Epub 2011 Oct 14. PLoS One. 2011. PMID: 22022466 Free PMC article.
-
Contributions of space-clamp errors to apparent time-dependent loss of Mg2+ block induced by NMDA.J Neurophysiol. 2017 Jul 1;118(1):532-543. doi: 10.1152/jn.00106.2017. Epub 2017 Mar 29. J Neurophysiol. 2017. PMID: 28356471 Free PMC article.
-
Beyond Activation: Characterizing Microglial Functional Phenotypes.Cells. 2021 Aug 28;10(9):2236. doi: 10.3390/cells10092236. Cells. 2021. PMID: 34571885 Free PMC article. Review.
-
Microglial activation by purines and pyrimidines.Glia. 2002 Nov;40(2):156-163. doi: 10.1002/glia.10150. Glia. 2002. PMID: 12379903 Review.
Cited by
-
Receptors on Microglia.Adv Neurobiol. 2024;37:83-121. doi: 10.1007/978-3-031-55529-9_6. Adv Neurobiol. 2024. PMID: 39207688 Review.
-
The formative role of microglia in stress-induced synaptic deficits and associated behavioral consequences.Neurosci Lett. 2019 Oct 15;711:134369. doi: 10.1016/j.neulet.2019.134369. Epub 2019 Aug 15. Neurosci Lett. 2019. PMID: 31422099 Free PMC article. Review.
-
Immune-vascular mural cell interactions: consequences for immune cell trafficking, cerebral blood flow, and the blood-brain barrier.Neurophotonics. 2022 Jul;9(3):031914. doi: 10.1117/1.NPh.9.3.031914. Epub 2022 May 14. Neurophotonics. 2022. PMID: 35581998 Free PMC article.
-
Pannexin-1 opening in neuronal edema causes cell death but also leads to protection via increased microglia contacts.Cell Rep. 2023 Oct 31;42(10):113128. doi: 10.1016/j.celrep.2023.113128. Epub 2023 Sep 23. Cell Rep. 2023. PMID: 37742194 Free PMC article.
-
Microglia: Lifelong patrolling immune cells of the brain.Prog Neurobiol. 2019 Aug;179:101614. doi: 10.1016/j.pneurobio.2019.04.003. Epub 2019 May 7. Prog Neurobiol. 2019. PMID: 31075285 Free PMC article. Review.
References
-
- Ambrosi C, Gassmann O, Pranskevich JN, Boassa D, Smock A, Wang J, Dahl G, Steinem C, Sosinsky GE. Pannexin1 and Pannexin2 channels show quaternary similarities to connexons and different oligomerization numbers from each other. J Biol Chem. 2010;285:24420–24431. doi: 10.1074/jbc.M110.115444. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
