Characterization of the influence of mediator complex in HIV-1 transcription
- PMID: 25100719
- PMCID: PMC4183804
- DOI: 10.1074/jbc.M114.570341
Characterization of the influence of mediator complex in HIV-1 transcription
Abstract
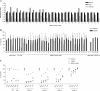
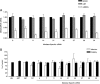
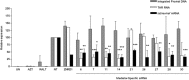
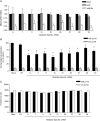
HIV-1 exploits multiple host proteins during infection. siRNA-based screenings have identified new proteins implicated in different pathways of the viral cycle that participate in a broad range of cellular functions. The human Mediator complex (MED) is composed of 28 elements and represents a fundamental component of the transcription machinery, interacting with the RNA polymerase II enzyme and regulating its ability to express genes. Here, we provide an evaluation of the MED activity on HIV replication. Knockdown of 9 out of 28 human MED proteins significantly impaired viral replication without affecting cell viability, including MED6, MED7, MED11, MED14, MED21, MED26, MED27, MED28, and MED30. Impairment of viral replication by MED subunits was at a post-integration step. Inhibition of early HIV transcripts was observed by siRNA-mediated knockdown of MED6, MED7, MED11, MED14, and MED28, specifically affecting the transcription of the nascent viral mRNA transactivation-responsive element. In addition, MED14 and MED30 were shown to have special relevance during the formation of unspliced viral transcripts (p < 0.0005). Knockdown of the selected MED factors compromised HIV transcription induced by Tat, with the strongest inhibitory effect shown by siMED6 and siMED14 cells. Co-immunoprecipitation experiments suggested physical interaction between MED14 and HIV-1 Tat protein. A better understanding of the mechanisms and factors controlling HIV-1 transcription is key to addressing the development of new strategies required to inhibit HIV replication or reactivate HIV-1 from the latent reservoirs.
Keywords: Host Factor; Human Immunodeficiency Virus (HIV); RNA Polymerase II; RNA Silencing; RNAP II; Tat; Transcription; Viral Transcription.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Ballana E., Esté J. A. (2013) Insights from host genomics into HIV infection and disease: identification of host targets for drug development. Antiviral Res. 100, 473–786 - PubMed
-
- Marcello A., Zoppé M., Giacca M. (2001) Multiple modes of transcriptional regulation by the HIV-1 Tat transactivator. IUBMB Life 51, 175–181 - PubMed
-
- Kao S. Y., Calman A. F., Luciw P. A., Peterlin B. M. (1987) Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330, 489–493 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

