The genomes, proteomes, and structures of three novel phages that infect the Bacillus cereus group and carry putative virulence factors
- PMID: 25100842
- PMCID: PMC4178739
- DOI: 10.1128/JVI.01364-14
The genomes, proteomes, and structures of three novel phages that infect the Bacillus cereus group and carry putative virulence factors
Abstract
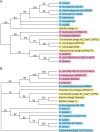
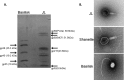
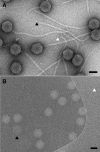
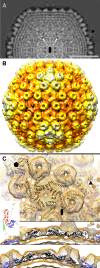
This article reports the results of studying three novel bacteriophages, JL, Shanette, and Basilisk, which infect the pathogen Bacillus cereus and carry genes that may contribute to its pathogenesis. We analyzed host range and superinfection ability, mapped their genomes, and characterized phage structure by mass spectrometry and transmission electron microscopy (TEM). The JL and Shanette genomes were 96% similar and contained 217 open reading frames (ORFs) and 220 ORFs, respectively, while Basilisk has an unrelated genome containing 138 ORFs. Mass spectrometry revealed 23 phage particle proteins for JL and 15 for Basilisk, while only 11 and 4, respectively, were predicted to be present by sequence analysis. Structural protein homology to well-characterized phages suggested that JL and Shanette were members of the family Myoviridae, which was confirmed by TEM. The third phage, Basilisk, was similar only to uncharacterized phages and is an unrelated siphovirus. Cryogenic electron microscopy of this novel phage revealed a T=9 icosahedral capsid structure with the major capsid protein (MCP) likely having the same fold as bacteriophage HK97 MCP despite the lack of sequence similarity. Several putative virulence factors were encoded by these phage genomes, including TerC and TerD involved in tellurium resistance. Host range analysis of all three phages supports genetic transfer of such factors within the B. cereus group, including B. cereus, B. anthracis, and B. thuringiensis. This study provides a basis for understanding these three phages and other related phages as well as their contributions to the pathogenicity of B. cereus group bacteria. Importance: The Bacillus cereus group of bacteria contains several human and plant pathogens, including B. cereus, B. anthracis, and B. thuringiensis. Phages are intimately linked to the evolution of their bacterial hosts and often provide virulence factors, making the study of B. cereus phages important to understanding the evolution of pathogenic strains. Herein we provide the results of detailed study of three novel B. cereus phages, two highly related myoviruses (JL and Shanette) and an unrelated siphovirus (Basilisk). The detailed characterization of host range and superinfection, together with results of genomic, proteomic, and structural analyses, reveal several putative virulence factors as well as the ability of these phages to infect different pathogenic species.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

