Specific argonautes selectively bind small RNAs derived from potato spindle tuber viroid and attenuate viroid accumulation in vivo
- PMID: 25100851
- PMCID: PMC4178711
- DOI: 10.1128/JVI.01404-14
Specific argonautes selectively bind small RNAs derived from potato spindle tuber viroid and attenuate viroid accumulation in vivo
Abstract
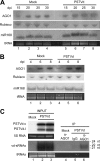
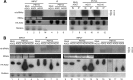
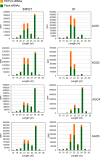
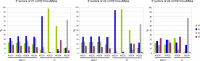
The identification of viroid-derived small RNAs (vd-sRNAs) of 21 to 24 nucleotides (nt) in plants infected by viroids (infectious non-protein-coding RNAs of just 250 to 400 nt) supports their targeting by Dicer-like enzymes, the first host RNA-silencing barrier. However, whether viroids, like RNA viruses, are also targeted by the RNA-induced silencing complex (RISC) remains controversial. At the RISC core is one Argonaute (AGO) protein that, guided by endogenous or viral sRNAs, targets complementary RNAs. To examine whether AGO proteins also load vd-sRNAs, leaves of Nicotiana benthamiana infected by potato spindle tuber viroid (PSTVd) were agroinfiltrated with plasmids expressing epitope-tagged versions of AGO1, AGO2, AGO3, AGO4, AGO5, AGO6, AGO7, AGO9, and AGO10 from Arabidopsis thaliana. Immunoprecipitation analyses of the agroinfiltrated halos revealed that all AGOs except AGO6, AGO7, and AGO10 associated with vd-sRNAs: AGO1, AGO2, and AGO3 preferentially with those of 21 and 22 nt, while AGO4, AGO5, and AGO9 additionally bound those of 24 nt. Deep-sequencing analyses showed that sorting of vd-sRNAs into AGO1, AGO2, AGO4, and AGO5 depended essentially on their 5'-terminal nucleotides, with the profiles of the corresponding AGO-loaded vd-sRNAs adopting specific hot spot distributions along the viroid genome. Furthermore, agroexpression of AGO1, AGO2, AGO4, and AGO5 on PSTVd-infected tissue attenuated the level of the genomic RNAs, suggesting that they, or their precursors, are RISC targeted. In contrast to RNA viruses, PSTVd infection of N. benthamiana did not affect miR168-mediated regulation of the endogenous AGO1, which loaded vd-sRNAs with specificity similar to that of its A. thaliana counterpart. Importance: To contain invaders, particularly RNA viruses, plants have evolved an RNA-silencing mechanism relying on the generation by Dicer-like (DCL) enzymes of virus-derived small RNAs of 21 to 24 nucleotides (nt) that load and guide Argonaute (AGO) proteins to target and repress viral RNA. Viroids, despite their minimal genomes (non-protein-coding RNAs of only 250 to 400 nt), infect and incite disease in plants. The accumulation in these plants of 21- to 24-nt viroid-derived small RNAs (vd-sRNAs) supports the notion that DCLs also target viroids but does not clarify whether vd-sRNAs activate one or more AGOs. Here, we show that in leaves of Nicotiana benthamiana infected by potato spindle tuber viroid, the endogenous AGO1 and distinct AGOs from Arabidopsis thaliana that were overexpressed were associated with vd-sRNAs displaying the same properties (5'-terminal nucleotide and size) previously established for endogenous and viral small RNAs. Overexpression of AGO1, AGO2, AGO4, and AGO5 attenuated viroid accumulation, supporting their role in antiviroid defense.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Degradome Analysis of Tomato and Nicotiana benthamiana Plants Infected with Potato Spindle Tuber Viroid.Int J Mol Sci. 2021 Apr 2;22(7):3725. doi: 10.3390/ijms22073725. Int J Mol Sci. 2021. PMID: 33918424 Free PMC article.
-
Silencing of transcription factor encoding gene StTCP23 by small RNAs derived from the virulence modulating region of potato spindle tuber viroid is associated with symptom development in potato.PLoS Pathog. 2019 Dec 2;15(12):e1008110. doi: 10.1371/journal.ppat.1008110. eCollection 2019 Dec. PLoS Pathog. 2019. PMID: 31790500 Free PMC article.
-
RNA-dependent RNA polymerase 6 delays accumulation and precludes meristem invasion of a viroid that replicates in the nucleus.J Virol. 2010 Mar;84(5):2477-89. doi: 10.1128/JVI.02336-09. Epub 2009 Dec 16. J Virol. 2010. PMID: 20015979 Free PMC article.
-
Viroid pathogenesis: a critical appraisal of the role of RNA silencing in triggering the initial molecular lesion.FEMS Microbiol Rev. 2020 May 1;44(3):386-398. doi: 10.1093/femsre/fuaa011. FEMS Microbiol Rev. 2020. PMID: 32379313 Review.
-
Viroids: how to infect a host and cause disease without encoding proteins.Biochimie. 2012 Jul;94(7):1474-80. doi: 10.1016/j.biochi.2012.02.020. Epub 2012 Feb 23. Biochimie. 2012. PMID: 22738729 Review.
Cited by
-
Replicating Potato spindle tuber viroid mediates de novo methylation of an intronic viroid sequence but no cleavage of the corresponding pre-mRNA.RNA Biol. 2015;12(3):268-75. doi: 10.1080/15476286.2015.1017216. RNA Biol. 2015. PMID: 25826660 Free PMC article.
-
High-Throughput Sequencing Analysis of Small RNAs Derived from Coleus Blumei Viroids.Viruses. 2019 Jul 5;11(7):619. doi: 10.3390/v11070619. Viruses. 2019. PMID: 31284471 Free PMC article.
-
Host factors against plant viruses.Mol Plant Pathol. 2019 Nov;20(11):1588-1601. doi: 10.1111/mpp.12851. Epub 2019 Jul 8. Mol Plant Pathol. 2019. PMID: 31286679 Free PMC article. Review.
-
Understanding Citrus Viroid Interactions: Experience and Prospects.Viruses. 2024 Apr 9;16(4):577. doi: 10.3390/v16040577. Viruses. 2024. PMID: 38675919 Free PMC article. Review.
-
Degradome Analysis of Tomato and Nicotiana benthamiana Plants Infected with Potato Spindle Tuber Viroid.Int J Mol Sci. 2021 Apr 2;22(7):3725. doi: 10.3390/ijms22073725. Int J Mol Sci. 2021. PMID: 33918424 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

