Temporal dynamics of fibrolytic and methanogenic rumen microorganisms during in situ incubation of switchgrass determined by 16S rRNA gene profiling
- PMID: 25101058
- PMCID: PMC4106096
- DOI: 10.3389/fmicb.2014.00307
Temporal dynamics of fibrolytic and methanogenic rumen microorganisms during in situ incubation of switchgrass determined by 16S rRNA gene profiling
Abstract
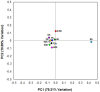
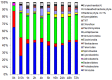
The rumen microbial ecosystem is known for its biomass-degrading and methane-producing phenotype. Fermentation of recalcitrant plant material, comprised of a multitude of interwoven fibers, necessitates the synergistic activity of diverse microbial taxonomic groups that inhabit the anaerobic rumen ecosystem. Although interspecies hydrogen (H2) transfer, a process during which bacterially generated H2 is transferred to methanogenic Archaea, has obtained significant attention over the last decades, the temporal variation of the different taxa involved in in situ biomass-degradation, H2 transfer and the methanogenesis process remains to be established. Here we investigated the temporal succession of microbial taxa and its effect on fiber composition during rumen incubation using 16S rRNA amplicon sequencing. Switchgrass filled nylon bags were placed in the rumen of a cannulated cow and collected at nine time points for DNA extraction and 16S pyrotag profiling. The microbial community colonizing the air-dried and non-incubated (0 h) switchgrass was dominated by members of the Bacilli (recruiting 63% of the pyrotag reads). During in situ incubation of the switchgrass, two major shifts in the community composition were observed: Bacilli were replaced within 30 min by members belonging to the Bacteroidia and Clostridia, which recruited 34 and 25% of the 16S rRNA reads generated, respectively. A second significant shift was observed after 16 h of rumen incubation, when members of the Spirochaetes and Fibrobacteria classes became more abundant in the fiber-adherent community. During the first 30 min of rumen incubation ~13% of the switchgrass dry matter was degraded, whereas little biomass degradation appeared to have occurred between 30 min and 4 h after the switchgrass was placed in the rumen. Interestingly, methanogenic members of the Euryarchaeota (i.e., Methanobacteria) increased up to 3-fold during this period of reduced biomass-degradation, with peak abundance just before rates of dry matter degradation increased again. We hypothesize that during this period microbial-mediated fibrolysis was temporarily inhibited until H2 was metabolized into CH4 by methanogens. Collectively, our results demonstrate the importance of inter-species interactions for the biomass-degrading and methane-producing phenotype of the rumen microbiome-both microbially facilitated processes with global significance.
Keywords: cellulolytic bacteria; interspecies H2 transfer; methanogenic archaea; microbe-microbe interactions; rumen microbiology.
Figures



Similar articles
-
Characterization and comparison of the temporal dynamics of ruminal bacterial microbiota colonizing rice straw and alfalfa hay within ruminants.J Dairy Sci. 2016 Dec;99(12):9668-9681. doi: 10.3168/jds.2016-11398. Epub 2016 Sep 28. J Dairy Sci. 2016. PMID: 27692708
-
3-Nitrooxypropanol supplementation had little effect on fiber degradation and microbial colonization of forage particles when evaluated using the in situ ruminal incubation technique.J Dairy Sci. 2020 Oct;103(10):8986-8997. doi: 10.3168/jds.2019-18077. Epub 2020 Aug 26. J Dairy Sci. 2020. PMID: 32861497
-
Structural and functional analysis of the active cow rumen's microbial community provides a catalogue of genes and microbes participating in the deconstruction of cardoon biomass.Biotechnol Biofuels Bioprod. 2024 Apr 8;17(1):53. doi: 10.1186/s13068-024-02495-4. Biotechnol Biofuels Bioprod. 2024. PMID: 38589938 Free PMC article.
-
Interactions between Anaerobic Fungi and Methanogens in the Rumen and Their Biotechnological Potential in Biogas Production from Lignocellulosic Materials.Microorganisms. 2021 Jan 17;9(1):190. doi: 10.3390/microorganisms9010190. Microorganisms. 2021. PMID: 33477342 Free PMC article. Review.
-
Upflow anaerobic sludge blanket reactor--a review.Indian J Environ Health. 2001 Apr;43(2):1-82. Indian J Environ Health. 2001. PMID: 12397675 Review.
Cited by
-
Conservation of Endophyte Bacterial Community Structure Across Two Panicum Grass Species.Front Microbiol. 2019 Sep 27;10:2181. doi: 10.3389/fmicb.2019.02181. eCollection 2019. Front Microbiol. 2019. PMID: 31611851 Free PMC article.
-
Temporal dynamics of in-situ fiber-adherent bacterial community under ruminal acidotic conditions determined by 16S rRNA gene profiling.PLoS One. 2017 Aug 1;12(8):e0182271. doi: 10.1371/journal.pone.0182271. eCollection 2017. PLoS One. 2017. PMID: 28763489 Free PMC article.
-
Using 'Omic Approaches to Compare Temporal Bacterial Colonization of Lolium perenne, Lotus corniculatus, and Trifolium pratense in the Rumen.Front Microbiol. 2018 Sep 19;9:2184. doi: 10.3389/fmicb.2018.02184. eCollection 2018. Front Microbiol. 2018. PMID: 30283417 Free PMC article.
-
Fibrolytic rumen bacteria of camel and sheep and their applications in the bioconversion of barley straw to soluble sugars for biofuel production.PLoS One. 2022 Jan 7;17(1):e0262304. doi: 10.1371/journal.pone.0262304. eCollection 2022. PLoS One. 2022. PMID: 34995335 Free PMC article.
-
Insights into the bacterial community and its temporal succession during the fermentation of wine grapes.Front Microbiol. 2015 Aug 18;6:809. doi: 10.3389/fmicb.2015.00809. eCollection 2015. Front Microbiol. 2015. PMID: 26347718 Free PMC article.
References
-
- Bowman J. G., Firkins J. L. (1993). Effects of forage species and particle size on bacterial cellulolytic activity and colonization in situ. J. Anim. Sci. 71, 1623–1633 - PubMed
-
- Coleman G. S. (1992). The rate of uptake and metabolism of starch grains and cellulose particles by Entodinium species, Eudiplodinium maggii, some other entodiniomorphid protozoa and natural protozoal populations taken from the ovine rumen. J. Appl. Bacteriol. 73, 507–513 10.1111/j.1365-2672.1992.tb05013.x - DOI - PubMed
-
- Corpe W. A. (1985). A method for detecting methylotrophic bacteria on solid-surfaces. J. Microbiol. Methods 3, 215–221 10.1016/0167-7012(85)90049-1 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

