Pattern Recognition and Signaling Mechanisms of RIG-I and MDA5
- PMID: 25101084
- PMCID: PMC4107945
- DOI: 10.3389/fimmu.2014.00342
Pattern Recognition and Signaling Mechanisms of RIG-I and MDA5
Abstract
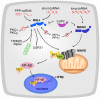
Most organisms rely on innate immune receptors to recognize conserved molecular structures from invading microbes. Two essential innate immune receptors, RIG-I and MDA5, detect viral double-stranded RNA in the cytoplasm. The inflammatory response triggered by these RIG-I-like receptors (RLRs) is one of the first and most important lines of defense against infection. RIG-I recognizes short RNA ligands with 5'-triphosphate caps. MDA5 recognizes long kilobase-scale genomic RNA and replication intermediates. Ligand binding induces conformational changes and oligomerization of RLRs that activate the signaling partner MAVS on the mitochondrial and peroxisomal membranes. This signaling process is under tight regulation, dependent on post-translational modifications of RIG-I and MDA5, and on regulatory proteins including unanchored ubiquitin chains and a third RLR, LGP2. Here, we review recent advances that have shifted the paradigm of RLR signaling away from the conventional linear signaling cascade. In the emerging RLR signaling model, large multimeric signaling platforms generate a highly cooperative, self-propagating, and context-dependent signal, which varies with the subcellular localization of the signaling platform.
Keywords: RecA-like DEAD-box (DExD/H-box) RNA helicase; amyloid-like aggregation; caspase recruitment domain; nucleic-acid sensor; pathogen-associated molecular pattern; prion-like switch; signal transduction; signalosome.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

