Repeat-mediated genetic and epigenetic changes at the FMR1 locus in the Fragile X-related disorders
- PMID: 25101111
- PMCID: PMC4101883
- DOI: 10.3389/fgene.2014.00226
Repeat-mediated genetic and epigenetic changes at the FMR1 locus in the Fragile X-related disorders
Abstract
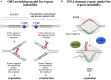
The Fragile X-related disorders are a group of genetic conditions that include the neurodegenerative disorder, Fragile X-associated tremor/ataxia syndrome (FXTAS), the fertility disorder, Fragile X-associated primary ovarian insufficiency (FXPOI) and the intellectual disability, Fragile X syndrome (FXS). The pathology in all these diseases is related to the number of CGG/CCG-repeats in the 5' UTR of the Fragile X mental retardation 1 (FMR1) gene. The repeats are prone to continuous expansion and the increase in repeat number has paradoxical effects on gene expression increasing transcription on mid-sized alleles and decreasing it on longer ones. In some cases the repeats can simultaneously both increase FMR1 mRNA production and decrease the levels of the FMR1 gene product, Fragile X mental retardation 1 protein (FMRP). Since FXTAS and FXPOI result from the deleterious consequences of the expression of elevated levels of FMR1 mRNA and FXS is caused by an FMRP deficiency, the clinical picture is turning out to be more complex than once appreciated. Added complications result from the fact that increasing repeat numbers make the alleles somatically unstable. Thus many individuals have a complex mixture of different sized alleles in different cells. Furthermore, it has become apparent that the eponymous fragile site, once thought to be no more than a useful diagnostic criterion, may have clinical consequences for females who inherit chromosomes that express this site. This review will cover what is currently known about the mechanisms responsible for repeat instability, for the repeat-mediated epigenetic changes that affect expression of the FMR1 gene, and for chromosome fragility. It will also touch on what current and future options are for ameliorating some of these effects.
Keywords: FX-associated primary ovarian insufficiency (FXPOI); FX-associated tremor/ataxia syndrome (FXTAS); Fragile X syndrome (FXS); Fragile X-related disorders; repeat expansion disease.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

