SCF(SLF)-mediated cytosolic degradation of S-RNase is required for cross-pollen compatibility in S-RNase-based self-incompatibility in Petunia hybrida
- PMID: 25101113
- PMCID: PMC4106197
- DOI: 10.3389/fgene.2014.00228
SCF(SLF)-mediated cytosolic degradation of S-RNase is required for cross-pollen compatibility in S-RNase-based self-incompatibility in Petunia hybrida
Abstract
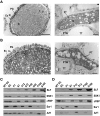
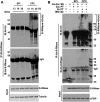
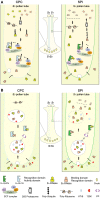
Many flowering plants adopt self-incompatibility (SI) to maintain their genetic diversity. In species of Solanaceae, Plantaginaceae, and Rosaceae, SI is genetically controlled by a single S-locus with multiple haplotypes. The S-locus has been shown to encode S-RNases expressed in pistil and multiple SLF (S-locus F-box) proteins in pollen controlling the female and male specificity of SI, respectively. S-RNases appear to function as a cytotoxin to reject self-pollen. In addition, SLFs have been shown to form SCF (SKP1/Cullin1/F-box) complexes to serve as putative E3 ubiquitin ligase to interact with S-RNases. Previously, two different mechanisms, the S-RNase degradation and the S-RNase compartmentalization, have been proposed as the restriction mechanisms of S-RNase cytotoxicity allowing compatible pollination. In this study, we have provided several lines of evidence in support of the S-RNase degradation mechanism by a combination of cellular, biochemical and molecular biology approaches. First, both immunogold labeling and subcellular fractionation assays showed that two key pollen SI factors, PhS3L-SLF1 and PhSSK1 (SLF-interacting SKP1-like1) from Petunia hybrida, a Solanaceous species, are co-localized in cytosols of both pollen grains and tubes. Second, PhS3L-RNases are mainly detected in the cytosols of both self and non-self-pollen tubes after pollination. Third, we found that PhS-RNases selectively interact with PhSLFs by yeast two-hybrid and co-immunoprecipitation assays. Fourth, S-RNases are specifically degraded in compatible pollen tubes by non-self SLF action. Taken together, our results demonstrate that SCF(SLF-mediated) non-self S-RNase degradation occurs in the cytosol of pollen tube through the ubiquitin/26S proteasome system serving as the major mechanism to neutralize S-RNase cytotoxicity during compatible pollination in P. hybrida.
Keywords: S-RNase localization; SCFSLF; cross-pollen compatibility; self-incompatibility; self-pollen incompatibility; ubiquitin/26S proteasome system.
Figures



Similar articles
-
Cullin1-P is an Essential Component of Non-Self Recognition System in Self-Incompatibility in Petunia.Plant Cell Physiol. 2016 Nov;57(11):2403-2416. doi: 10.1093/pcp/pcw152. Epub 2016 Aug 26. Plant Cell Physiol. 2016. PMID: 27565207
-
CRISPR/Cas9-mediated knockout of PiSSK1 reveals essential role of S-locus F-box protein-containing SCF complexes in recognition of non-self S-RNases during cross-compatible pollination in self-incompatible Petunia inflata.Plant Reprod. 2018 Jun;31(2):129-143. doi: 10.1007/s00497-017-0314-1. Epub 2017 Nov 30. Plant Reprod. 2018. PMID: 29192328
-
Ubiquitin-proteasome-mediated degradation of S-RNase in a solanaceous cross-compatibility reaction.Plant J. 2014 Jun;78(6):1014-21. doi: 10.1111/tpj.12528. Epub 2014 May 21. Plant J. 2014. PMID: 24689760
-
S-RNase-based self-incompatibility in Petunia inflata.Ann Bot. 2011 Sep;108(4):637-46. doi: 10.1093/aob/mcq253. Epub 2010 Dec 30. Ann Bot. 2011. PMID: 21193481 Free PMC article. Review.
-
Biochemical models for S-RNase-based self-incompatibility.Mol Plant. 2008 Jul;1(4):575-85. doi: 10.1093/mp/ssn032. Epub 2008 Jun 26. Mol Plant. 2008. PMID: 19825563 Review.
Cited by
-
Aminooxyacetic acid (АОА), inhibitor of 1-aminocyclopropane-1-carboxilic acid (AСС) synthesis, suppresses self-incompatibility-induced programmed cell death in self-incompatible Petunia hybrida L. pollen tubes.Protoplasma. 2020 Jan;257(1):213-227. doi: 10.1007/s00709-019-01430-x. Epub 2019 Aug 13. Protoplasma. 2020. PMID: 31410589
-
Recognition of S-RNases by an S locus F-box like protein and an S haplotype-specific F-box like protein in the Prunus-specific self-incompatibility system.Plant Mol Biol. 2019 Jul;100(4-5):367-378. doi: 10.1007/s11103-019-00860-8. Epub 2019 Apr 1. Plant Mol Biol. 2019. PMID: 30937702
-
SLFL Genes Participate in the Ubiquitination and Degradation Reaction of S-RNase in Self-compatible Peach.Front Plant Sci. 2018 Feb 22;9:227. doi: 10.3389/fpls.2018.00227. eCollection 2018. Front Plant Sci. 2018. PMID: 29520292 Free PMC article.
-
Comparative Proteomics Analyses of Pollination Response in Endangered Orchid Species Dendrobium Chrysanthum.Int J Mol Sci. 2017 Nov 23;18(12):2496. doi: 10.3390/ijms18122496. Int J Mol Sci. 2017. PMID: 29168730 Free PMC article.
-
Molecular mechanisms and genetic regulation of self-incompatibility in flowering plants: implications for crop improvement and evolutionary biology.Plant Mol Biol. 2025 Jun 25;115(4):76. doi: 10.1007/s11103-025-01610-9. Plant Mol Biol. 2025. PMID: 40560319 Review.
References
-
- Arrese E. L., Gazard J. L., Flowers M. T., Soulages J. L., Wells M. A. (2001). Diacylglycerol transport in the insect fat body: evidence of involvement of lipid droplets and the cytosolic fraction. Lipid Res. 42, 225–234 - PubMed
-
- Busot G. Y., McClure B., Ibarra-Sánchez C. P., Jiménez-Durán K., Vázquez-Santana S., Cruz-García F. (2008). Pollination in Nicotiana alata stimulates synthesis and transfer to the stigmatic surface of NaStEP, a vacuolar Kunitz proteinase inhibitor homologue. J. Exp. Bot. 59, 3187–3201 10.1093/jxb/ern175 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

