Comprehensive identification of single nucleotide polymorphisms associated with beta-lactam resistance within pneumococcal mosaic genes
- PMID: 25101644
- PMCID: PMC4125147
- DOI: 10.1371/journal.pgen.1004547
Comprehensive identification of single nucleotide polymorphisms associated with beta-lactam resistance within pneumococcal mosaic genes
Abstract
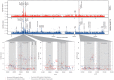
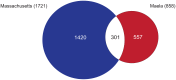
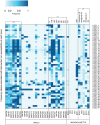
Traditional genetic association studies are very difficult in bacteria, as the generally limited recombination leads to large linked haplotype blocks, confounding the identification of causative variants. Beta-lactam antibiotic resistance in Streptococcus pneumoniae arises readily as the bacteria can quickly incorporate DNA fragments encompassing variants that make the transformed strains resistant. However, the causative mutations themselves are embedded within larger recombined blocks, and previous studies have only analysed a limited number of isolates, leading to the description of "mosaic genes" as being responsible for resistance. By comparing a large number of genomes of beta-lactam susceptible and non-susceptible strains, the high frequency of recombination should break up these haplotype blocks and allow the use of genetic association approaches to identify individual causative variants. Here, we performed a genome-wide association study to identify single nucleotide polymorphisms (SNPs) and indels that could confer beta-lactam non-susceptibility using 3,085 Thai and 616 USA pneumococcal isolates as independent datasets for the variant discovery. The large sample sizes allowed us to narrow the source of beta-lactam non-susceptibility from long recombinant fragments down to much smaller loci comprised of discrete or linked SNPs. While some loci appear to be universal resistance determinants, contributing equally to non-susceptibility for at least two classes of beta-lactam antibiotics, some play a larger role in resistance to particular antibiotics. All of the identified loci have a highly non-uniform distribution in the populations. They are enriched not only in vaccine-targeted, but also non-vaccine-targeted lineages, which may raise clinical concerns. Identification of single nucleotide polymorphisms underlying resistance will be essential for future use of genome sequencing to predict antibiotic sensitivity in clinical microbiology.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Molecular Characterization of β-Lactam Resistant Streptococcus pneumoniae.Clin Lab. 2020 Jan 1;66(1). doi: 10.7754/Clin.Lab.2019.190529. Clin Lab. 2020. PMID: 32013352
-
[A mass-spectrometric analysis of genetic markers of S. pneumoniae resistance to beta-lactam antibiotics].Mol Gen Mikrobiol Virusol. 2010;(3):16-25. Mol Gen Mikrobiol Virusol. 2010. PMID: 20882772 Russian.
-
Commensal streptococci serve as a reservoir for β-lactam resistance genes in Streptococcus pneumoniae.Antimicrob Agents Chemother. 2015;59(6):3529-40. doi: 10.1128/AAC.00429-15. Epub 2015 Apr 6. Antimicrob Agents Chemother. 2015. PMID: 25845880 Free PMC article.
-
Molecular mechanisms of β-lactam resistance in Streptococcus pneumoniae.Future Microbiol. 2012 Mar;7(3):395-410. doi: 10.2217/fmb.12.2. Future Microbiol. 2012. PMID: 22393892 Review.
-
[Beta-lactam and macrolide resistance in Streptococcus pneumoniae].Nihon Rinsho. 2001 Apr;59(4):681-6. Nihon Rinsho. 2001. PMID: 11304989 Review. Japanese.
Cited by
-
β-Lactam Resistance Mechanisms: Gram-Positive Bacteria and Mycobacterium tuberculosis.Cold Spring Harb Perspect Med. 2016 May 2;6(5):a025221. doi: 10.1101/cshperspect.a025221. Cold Spring Harb Perspect Med. 2016. PMID: 27091943 Free PMC article. Review.
-
Convergent impact of vaccination and antibiotic pressures on pneumococcal populations.Cell Chem Biol. 2024 Feb 15;31(2):195-206. doi: 10.1016/j.chembiol.2023.11.003. Epub 2023 Dec 4. Cell Chem Biol. 2024. PMID: 38052216 Free PMC article. Review.
-
Stem Region of tRNA Genes Favors Transition Substitution Towards Keto Bases in Bacteria.J Mol Evol. 2022 Feb;90(1):114-123. doi: 10.1007/s00239-021-10045-x. Epub 2022 Jan 27. J Mol Evol. 2022. PMID: 35084523
-
Antimicrobial susceptibility profile & resistance mechanisms of Global Antimicrobial Resistance Surveillance System (GLASS) priority pathogens from India.Indian J Med Res. 2019 Feb;149(2):87-96. doi: 10.4103/ijmr.IJMR_214_18. Indian J Med Res. 2019. PMID: 31219073 Free PMC article. Review.
-
Streptococcus suis serotype 9 in Italy: genomic insights into high-risk clones with emerging resistance to penicillin.J Antimicrob Chemother. 2024 Feb 1;79(2):403-411. doi: 10.1093/jac/dkad395. J Antimicrob Chemother. 2024. PMID: 38153239 Free PMC article.
References
-
- Hakenbeck R, Bruckner R, Denapaite D, Maurer P (2012) Molecular mechanisms of beta-lactam resistance in Streptococcus pneumoniae. Future microbiology 7: 395–410. - PubMed
-
- Crisostomo MI, Vollmer W, Kharat AS, Inhulsen S, Gehre F, et al. (2006) Attenuation of penicillin resistance in a peptidoglycan O-acetyl transferase mutant of Streptococcus pneumoniae. Molecular microbiology 61: 1497–1509. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

