Evidence for divisome localization mechanisms independent of the Min system and SlmA in Escherichia coli
- PMID: 25101671
- PMCID: PMC4125044
- DOI: 10.1371/journal.pgen.1004504
Evidence for divisome localization mechanisms independent of the Min system and SlmA in Escherichia coli
Abstract
Cell division in Escherichia coli starts with assembly of FtsZ protofilaments into a ring-like structure, the Z-ring. Positioning of the Z-ring at midcell is thought to be coordinated by two regulatory systems, nucleoid occlusion and the Min system. In E. coli, nucleoid occlusion is mediated by the SlmA proteins. Here, we address the question of whether there are additional positioning systems that are capable of localizing the E. coli divisome with respect to the cell center. Using quantitative fluorescence imaging we show that slow growing cells lacking functional Min and SlmA nucleoid occlusion systems continue to divide preferentially at midcell. We find that the initial Z-ring assembly occurs over the center of the nucleoid instead of nucleoid-free regions under these conditions. We determine that Z-ring formation begins shortly after the arrival of the Ter macrodomain at the nucleoid center. Removal of either the MatP, ZapB, or ZapA proteins significantly affects the accuracy and precision of Z-ring positioning relative to the nucleoid center in these cells in accordance with the idea that these proteins link the Ter macrodomain and the Z-ring. Interestingly, even in the absence of Min, SlmA, and the putative Ter macrodomain - Z-ring link, there remains a weak midcell positioning bias for the Z-ring. Our work demonstrates that additional Z-ring localization systems are present in E. coli than are known currently. In particular, we identify that the Ter macrodomain acts as a landmark for the Z-ring in the presence of MatP, ZapB and ZapA proteins.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

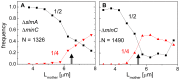
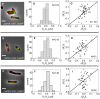
 . Data are shown only for cells with a single nucleoid. (D) ΔXz vs. ΔXn for the parental strain (strain JMBW5). (E), (F) Distribution of distances between the Z-ring center and nucleoid center for ΔslmA Δmin strain and parental strain, respectively. Data for central Z-rings are shown. (G), (H) ΔXz vs. ΔXn for cells that show a Z-ring over a compact nucleoid in ΔslmA Δmin and in parental strain, respectively.
. Data are shown only for cells with a single nucleoid. (D) ΔXz vs. ΔXn for the parental strain (strain JMBW5). (E), (F) Distribution of distances between the Z-ring center and nucleoid center for ΔslmA Δmin strain and parental strain, respectively. Data for central Z-rings are shown. (G), (H) ΔXz vs. ΔXn for cells that show a Z-ring over a compact nucleoid in ΔslmA Δmin and in parental strain, respectively.
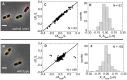
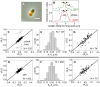
 . Only cells with a single MatP focus are analyzed. (D) ΔXz vs ΔXMatP for wild type cells. (E), (F) Distribution of distances between the Z-ring and the MatP focus along the long axes of the cell for ΔslmA Δmin and wild type cells, respectively.
. Only cells with a single MatP focus are analyzed. (D) ΔXz vs ΔXMatP for wild type cells. (E), (F) Distribution of distances between the Z-ring and the MatP focus along the long axes of the cell for ΔslmA Δmin and wild type cells, respectively.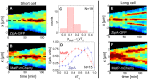
 .
.References
-
- Bi E, Lutkenhaus J (1991) FtsZ ring structure associated with division in Escherichia coli . Nature 354: 161–164. - PubMed
-
- Lutkenhaus J (2007) Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem 76: 539–562. - PubMed
-
- Adams DW, Errington J (2009) Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol 7: 642–653. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

