Playing RNase P evolution: swapping the RNA catalyst for a protein reveals functional uniformity of highly divergent enzyme forms
- PMID: 25101763
- PMCID: PMC4125048
- DOI: 10.1371/journal.pgen.1004506
Playing RNase P evolution: swapping the RNA catalyst for a protein reveals functional uniformity of highly divergent enzyme forms
Abstract
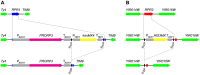
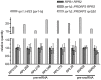
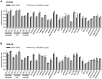
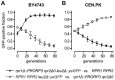
The RNase P family is a diverse group of endonucleases responsible for the removal of 5' extensions from tRNA precursors. The diversity of enzyme forms finds its extremes in the eukaryal nucleus where RNA-based catalysis by complex ribonucleoproteins in some organisms contrasts with single-polypeptide enzymes in others. Such structural contrast suggests associated functional differences, and the complexity of the ribonucleoprotein was indeed proposed to broaden the enzyme's functionality beyond tRNA processing. To explore functional overlap and differences between most divergent forms of RNase P, we replaced the nuclear RNase P of Saccharomyces cerevisiae, a 10-subunit ribonucleoprotein, with Arabidopsis thaliana PRORP3, a single monomeric protein. Surprisingly, the RNase P-swapped yeast strains were viable, displayed essentially unimpaired growth under a wide variety of conditions, and, in a certain genetic background, their fitness even slightly exceeded that of the wild type. The molecular analysis of the RNase P-swapped strains showed a minor disturbance in tRNA metabolism, but did not point to any RNase P substrates or functions beyond that. Altogether, these results indicate the full functional exchangeability of the highly dissimilar enzymes. Our study thereby establishes the RNase P family, with its combination of structural diversity and functional uniformity, as an extreme case of convergent evolution. It moreover suggests that the apparently gratuitous complexity of some RNase P forms is the result of constructive neutral evolution rather than reflecting increased functional versatility.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Distribution of Ribonucleoprotein and Protein-Only RNase P in Eukarya.Mol Biol Evol. 2015 Dec;32(12):3186-93. doi: 10.1093/molbev/msv187. Epub 2015 Sep 3. Mol Biol Evol. 2015. PMID: 26341299
-
Substrate recognition and cleavage-site selection by a single-subunit protein-only RNase P.Nucleic Acids Res. 2016 Mar 18;44(5):2323-36. doi: 10.1093/nar/gkw080. Epub 2016 Feb 20. Nucleic Acids Res. 2016. PMID: 26896801 Free PMC article.
-
PRORP proteins support RNase P activity in both organelles and the nucleus in Arabidopsis.Genes Dev. 2012 May 15;26(10):1022-7. doi: 10.1101/gad.189514.112. Epub 2012 May 1. Genes Dev. 2012. PMID: 22549728 Free PMC article.
-
Of P and Z: mitochondrial tRNA processing enzymes.Biochim Biophys Acta. 2012 Sep-Oct;1819(9-10):1017-26. doi: 10.1016/j.bbagrm.2011.11.003. Epub 2011 Nov 23. Biochim Biophys Acta. 2012. PMID: 22137969 Free PMC article. Review.
-
Multiple structural flavors of RNase P in precursor tRNA processing.Wiley Interdiscip Rev RNA. 2024 Mar-Apr;15(2):e1835. doi: 10.1002/wrna.1835. Wiley Interdiscip Rev RNA. 2024. PMID: 38479802 Review.
Cited by
-
The Constructive Neutral Evolution of Behaviour.Ecol Evol. 2025 Jul 10;15(7):e71736. doi: 10.1002/ece3.71736. eCollection 2025 Jul. Ecol Evol. 2025. PMID: 40641486 Free PMC article.
-
Essential is not irreplaceable: fitness dynamics of experimental E. coli RNase P RNA heterologous replacement.J Mol Evol. 2014 Oct;79(3-4):143-52. doi: 10.1007/s00239-014-9646-8. Epub 2014 Sep 30. J Mol Evol. 2014. PMID: 25266807
-
Auto-inhibitory Mechanism of the Human Mitochondrial RNase P Protein Complex.Sci Rep. 2015 Apr 30;5:9878. doi: 10.1038/srep09878. Sci Rep. 2015. PMID: 25928769 Free PMC article.
-
Mechanistic and Structural Studies of Protein-Only RNase P Compared to Ribonucleoproteins Reveal the Two Faces of the Same Enzymatic Activity.Biomolecules. 2016 Jun 24;6(3):30. doi: 10.3390/biom6030030. Biomolecules. 2016. PMID: 27348014 Free PMC article. Review.
-
Protein-only RNase P function in Escherichia coli: viability, processing defects and differences between PRORP isoenzymes.Nucleic Acids Res. 2017 Jul 7;45(12):7441-7454. doi: 10.1093/nar/gkx405. Nucleic Acids Res. 2017. PMID: 28499021 Free PMC article.
References
-
- Liu F, Altman S, editors (2010) Ribonuclease P. New York: Springer. 283 p.
-
- Hartmann RK, Göβringer M, Späth B, Fischer S, Marchfelder A (2009) The Making of tRNAs and More - RNase P and tRNase Z. Prog Nucleic Acid Res Mol Biol. 85: 319–368. - PubMed
-
- Ellis JC, Brown JW (2010) The evolution of RNase P and its RNA. In: Liu F, Altman S, editors. Ribonuclease P. New York: Springer. pp. 17–40.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

