Response regulator heterodimer formation controls a key stage in Streptomyces development
- PMID: 25101778
- PMCID: PMC4125116
- DOI: 10.1371/journal.pgen.1004554
Response regulator heterodimer formation controls a key stage in Streptomyces development
Abstract
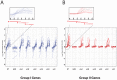
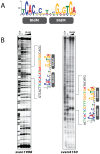
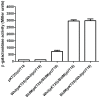
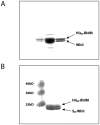
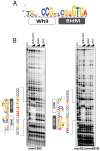
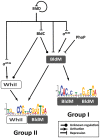
The orphan, atypical response regulators BldM and WhiI each play critical roles in Streptomyces differentiation. BldM is required for the formation of aerial hyphae, and WhiI is required for the differentiation of these reproductive structures into mature spores. To gain insight into BldM function, we defined the genome-wide BldM regulon using ChIP-Seq and transcriptional profiling. BldM target genes clustered into two groups based on their whi gene dependency. Expression of Group I genes depended on bldM but was independent of all the whi genes, and biochemical experiments showed that Group I promoters were controlled by a BldM homodimer. In contrast, Group II genes were expressed later than Group I genes and their expression depended not only on bldM but also on whiI and whiG (encoding the sigma factor that activates whiI). Additional ChIP-Seq analysis showed that BldM Group II genes were also direct targets of WhiI and that in vivo binding of WhiI to these promoters depended on BldM and vice versa. We go on to demonstrate that BldM and WhiI form a functional heterodimer that controls Group II promoters, serving to integrate signals from two distinct developmental pathways. The BldM-WhiI system thus exemplifies the potential of response regulator heterodimer formation as a mechanism to expand the signaling capabilities of bacterial cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Elliot MA, Buttner MJ, Nodwell JR (2008) Multicellular Development in Streptomyces In: Whitworth DE, editor. Myxobacteria: Multicellularity and Differentiation. Washington D.C.: ASM Press. pp. 419–438.
-
- Flärdh K, Buttner MJ (2009) Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol 7: 36–49. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

