Reduction of systematic bias in transcriptome data from human peripheral blood mononuclear cells for transportation and biobanking
- PMID: 25101803
- PMCID: PMC4125218
- DOI: 10.1371/journal.pone.0104283
Reduction of systematic bias in transcriptome data from human peripheral blood mononuclear cells for transportation and biobanking
Abstract
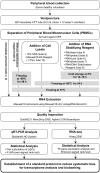
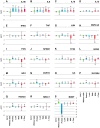
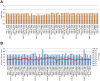
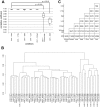
Transportation of samples is essential for large-scale biobank projects. However, RNA degradation during pre-analytical operations prior to transportation can cause systematic bias in transcriptome data, which may prevent subsequent biomarker identification. Therefore, to collect high-quality biobank samples for expression analysis, specimens must be transported under stable conditions. In this study, we examined the effectiveness of RNA-stabilizing reagents to prevent RNA degradation during pre-analytical operations with an emphasis on RNA from peripheral blood mononuclear cells (PBMCs) to establish a protocol for reducing systematic bias. To this end, we obtained PBMCs from 11 healthy volunteers and analyzed the purity, yield, and integrity of extracted RNA after performing pre-analytical operations for freezing PBMCs at -80°C. We randomly chose 7 samples from 11 samples individually, and systematic bias in expression levels was examined by real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR), RNA sequencing (RNA-Seq) experiments and data analysis. Our data demonstrated that omission of stabilizing reagents significantly lowered RNA integrity, suggesting substantial degradation of RNA molecules due to pre-analytical freezing. qRT-PCR experiments for 19 selected transcripts revealed systematic bias in the expression levels of five transcripts. RNA-Seq for 25,223 transcripts also suggested that about 40% of transcripts were systematically biased. These results indicated that appropriate reduction in systematic bias is essential in protocols for collection of RNA from PBMCs for large-scale biobank projects. Among the seven commercially available stabilizing reagents examined in this study, qRT-PCR and RNA-Seq experiments consistently suggested that RNALock, RNA/DNA Stabilization Reagent for Blood and Bone Marrow, and 1-Thioglycerol/Homogenization solution could reduce systematic bias. On the basis of the results of this study, we established a protocol to reduce systematic bias in the expression levels of RNA transcripts isolated from PBMCs. We believe that these data provide a novel methodology for collection of high-quality RNA from PBMCs for biobank researchers.
Conflict of interest statement
Figures

Similar articles
-
Investigating gene expression profiles of whole blood and peripheral blood mononuclear cells using multiple collection and processing methods.PLoS One. 2019 Dec 6;14(12):e0225137. doi: 10.1371/journal.pone.0225137. eCollection 2019. PLoS One. 2019. PMID: 31809517 Free PMC article.
-
Quality assessment on the long-term cryopreservation and nucleic acids extraction processes implemented in the andalusian public biobank.Cell Tissue Bank. 2019 Jun;20(2):255-265. doi: 10.1007/s10561-019-09764-9. Epub 2019 Mar 22. Cell Tissue Bank. 2019. PMID: 30903409
-
Effects of blood transportation on human peripheral mononuclear cell yield, phenotype and function: implications for immune cell biobanking.PLoS One. 2014 Dec 26;9(12):e115920. doi: 10.1371/journal.pone.0115920. eCollection 2014. PLoS One. 2014. PMID: 25541968 Free PMC article.
-
Impact of preanalytical handling and timing for peripheral blood mononuclear cells isolation and RNA studies: the experience of the Interinstitutional Multidisciplinary BioBank (BioBIM).Int J Biol Markers. 2012 Jul 19;27(2):e90-8. doi: 10.5301/JBM.2012.9235. Int J Biol Markers. 2012. PMID: 22562396
-
The circular RNA of peripheral blood mononuclear cells: Hsa_circ_0005836 as a new diagnostic biomarker and therapeutic target of active pulmonary tuberculosis.Mol Immunol. 2017 Oct;90:264-272. doi: 10.1016/j.molimm.2017.08.008. Epub 2017 Aug 30. Mol Immunol. 2017. PMID: 28846924
Cited by
-
Multiple freeze-thaw cycles lead to a loss of consistency in poly(A)-enriched RNA sequencing.BMC Genomics. 2021 Jan 21;22(1):69. doi: 10.1186/s12864-021-07381-z. BMC Genomics. 2021. PMID: 33478392 Free PMC article.
-
Impact of RNA integrity and blood sample storage conditions on the gene expression analysis.Onco Targets Ther. 2018 Jun 20;11:3573-3581. doi: 10.2147/OTT.S158868. eCollection 2018. Onco Targets Ther. 2018. PMID: 29950862 Free PMC article.
-
Genome-wide identification of inter-individually variable DNA methylation sites improves the efficacy of epigenetic association studies.NPJ Genom Med. 2017 Apr 13;2:11. doi: 10.1038/s41525-017-0016-5. eCollection 2017. NPJ Genom Med. 2017. PMID: 29263827 Free PMC article.
-
DNA Methylation Abnormalities and Altered Whole Transcriptome Profiles after Switching from Combustible Tobacco Smoking to Heated Tobacco Products.Cancer Epidemiol Biomarkers Prev. 2022 Jan;31(1):269-279. doi: 10.1158/1055-9965.EPI-21-0444. Epub 2021 Nov 2. Cancer Epidemiol Biomarkers Prev. 2022. PMID: 34728466 Free PMC article.
-
Optimized protocol for the extraction of RNA and DNA from frozen whole blood sample stored in a single EDTA tube.Sci Rep. 2021 Aug 23;11(1):17075. doi: 10.1038/s41598-021-96567-2. Sci Rep. 2021. PMID: 34426633 Free PMC article.
References
-
- Nakamura M, Tanaka F, Nakajima S, Honma M, Sakai T, et al. (2012) Comparison of the incidence of acute decompensated heart failure before and after the major tsunami in Northeast Japan. Am J Cardiol 110: 1856–1860. - PubMed
-
- Omama S, Yoshida Y, Ogasawara K, Ogawa A, Ishibashi Y, et al. (2013) Influence of the great East Japan earthquake and tsunami 2011 on occurrence of cerebrovascular diseases in Iwate, Japan. Stroke 44: 1518–1524. - PubMed
-
- Aoki T, Fukumoto Y, Yasuda S, Sakata Y, Ito K, et al. (2012) The Great East Japan Earthquake Disaster and cardiovascular diseases. Eur Heart J 33: 2796–2803. - PubMed
-
- Steinberg K, Beck J, Nickerson D, Garcia-Closas M, Gallagher M, et al. (2002) DNA banking for epidemiologic studies: a review of current practices. Epidemiology 13: 246–254. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

