ATG8 localization in apicomplexan parasites: apicoplast and more?
- PMID: 25102412
- PMCID: PMC4206529
- DOI: 10.4161/auto.32183
ATG8 localization in apicomplexan parasites: apicoplast and more?
Abstract
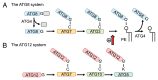
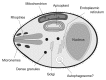
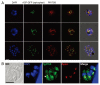
The ATG genes are highly conserved in eukaryotes including yeasts, plants, and mammals. However, these genes appear to be only partially present in most protists. Recent studies demonstrated that, in the apicomplexan parasites Plasmodium (malaria parasites) and Toxoplasma, ATG8 localizes to the apicoplast, a unique nonphotosynthetic plastid with 4 limiting membranes. In contrast to this established localization, it remains unclear whether these parasites can induce canonical macroautophagy and if ATG8 localizes to autophagosomes. Furthermore, the molecular function of ATG8 in its novel workplace, the apicoplast, is totally unknown. Here, we review recent studies on ATG8 in Plasmodium and Toxoplasma, summarize both consensus and controversial findings, and discuss its potential role in these parasites.
Keywords: ATG8; apicoplast; autophagy; plasmodium; toxoplasma.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
