Identification of copy number variants from exome sequence data
- PMID: 25102989
- PMCID: PMC4132917
- DOI: 10.1186/1471-2164-15-661
Identification of copy number variants from exome sequence data
Abstract
Background: With advances in next generation sequencing technologies and genomic capture techniques, exome sequencing has become a cost-effective approach for mutation detection in genetic diseases. However, computational prediction of copy number variants (CNVs) from exome sequence data is a challenging task. Whilst numerous programs are available, they have different sensitivities, and have low sensitivity to detect smaller CNVs (1-4 exons). Additionally, exonic CNV discovery using standard aCGH has limitations due to the low probe density over exonic regions. The goal of our study was to develop a protocol to detect exonic CNVs (including shorter CNVs that cover 1-4 exons), combining computational prediction algorithms and a high-resolution custom CGH array.
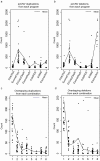
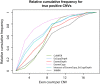
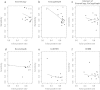
Results: We used six published CNV prediction programs (ExomeCNV, CONTRA, ExomeCopy, ExomeDepth, CoNIFER, XHMM) and an in-house modification to ExomeCopy and ExomeDepth (ExCopyDepth) for computational CNV prediction on 30 exomes from the 1000 genomes project and 9 exomes from primary immunodeficiency patients. CNV predictions were tested using a custom CGH array designed to capture all exons (exaCGH). After this validation, we next evaluated the computational prediction of shorter CNVs. ExomeCopy and the in-house modified algorithm, ExCopyDepth, showed the highest capability in detecting shorter CNVs. Finally, the performance of each computational program was assessed by calculating the sensitivity and false positive rate.
Conclusions: In this paper, we assessed the ability of 6 computational programs to predict CNVs, focussing on short (1-4 exon) CNVs. We also tested these predictions using a custom array targeting exons. Based on these results, we propose a protocol to identify and confirm shorter exonic CNVs combining computational prediction algorithms and custom aCGH experiments.
Figures

References
-
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. - DOI - PMC - PubMed
-
- De Ligt J, Boone PM, Pfundt R, Vissers LELM, Richmond T, Geoghegan J, O’Moore K, de Leeuw N, Shaw C, Brunner HG, Lupski JR, Veltman JA, Hehir-Kwa JY. Detection of clinically relevant copy number variants with whole-exome sequencing. Hum Mutat. 2013;34:1439–1448. doi: 10.1002/humu.22387. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

