The Caenorhabditis elegans pericentriolar material components SPD-2 and SPD-5 are monomeric in the cytoplasm before incorporation into the PCM matrix
- PMID: 25103243
- PMCID: PMC4230587
- DOI: 10.1091/mbc.E13-09-0514
The Caenorhabditis elegans pericentriolar material components SPD-2 and SPD-5 are monomeric in the cytoplasm before incorporation into the PCM matrix
Abstract
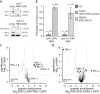
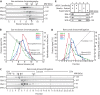
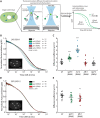
Centrosomes are the main microtubule-organizing centers in animal cells. Centrosomes consist of a pair of centrioles surrounded by a matrix of pericentriolar material (PCM) that assembles from cytoplasmic components. In Caenorhabditis elegans embryos, interactions between the coiled-coil proteins SPD-5 and SPD-2 and the kinase PLK-1 are critical for PCM assembly. However, it is not known whether these interactions promote the formation of cytoplasmic complexes that are added to the PCM or whether the components interact only during incorporation into the PCM matrix. Here we address this problem by using a combination of live-cell fluorescence correlation spectroscopy, mass spectrometry, and hydrodynamic techniques to investigate the native state of PCM components in the cytoplasm. We show that SPD-2 is monomeric, and neither SPD-2 nor SPD-5 exists in complex with PLK-1. SPD-5 exists mostly as a monomer but also forms complexes with the PP2A-regulatory proteins RSA-1 and RSA-2, which are required for microtubule organization at centrosomes. These results suggest that the interactions between SPD-2, SPD-5, and PLK-1 do not result in formation of cytoplasmic complexes, but instead occur in the context of PCM assembly.
© 2014 Wueseke et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures
References
-
- Bacia K, Kim SA, Schwille P. Fluorescence cross-correlation spectroscopy in living cells. Nat Methods. 2006;3:83–89. - PubMed
-
- Bacia K, Schwille P. Practical guidelines for dual-color fluorescence cross-correlation spectroscopy. Nat Protoc. 2007;2:2842–2856. - PubMed
-
- Baudendistel N, Müller G, Waldeck W. 2005. Two-hybrid fluorescence cross-correlation spectroscopy detects protein–protein interactions in vivo Chemphyschem 6, 984–990. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

