Argonaute CLIP-Seq reveals miRNA targetome diversity across tissue types
- PMID: 25103560
- PMCID: PMC4894423
- DOI: 10.1038/srep05947
Argonaute CLIP-Seq reveals miRNA targetome diversity across tissue types
Abstract
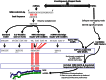
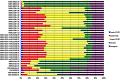
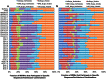
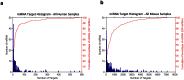
To date, analyses of individual targets have provided evidence of a miRNA targetome that extends beyond the boundaries of messenger RNAs (mRNAs) and can involve non-Watson-Crick base pairing in the miRNA seed region. Here we report our findings from analyzing 34 Argonaute HITS-CLIP datasets from several human and mouse cell types. Investigation of the architectural (i.e. bulge vs. contiguous pairs) and sequence (Watson-Crick vs. G:U pairs) preferences for human and mouse miRNAs revealed that many heteroduplexes are "non-canonical" i.e. their seed region comprises G:U and bulge combinations. The genomic distribution of miRNA targets differed distinctly across cell types but remained congruent across biological replicates of the same cell type. For some cell types intergenic and intronic targets were more frequent whereas in other cell types mRNA targets prevailed. The findings suggest an expanded model of miRNA targeting that is more frequent than the standard model currently in use. Lastly, our analyses of data from different cell types and laboratories revealed consistent Ago-loaded miRNA profiles across replicates whereas, unexpectedly, the Ago-loaded targets exhibited a much more dynamic behavior across biological replicates.
Figures


References
-
- Wightman B., Ha I. & Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862 (1993). - PubMed
-
- Lee R. C., Feinbaum R. L. & Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993). - PubMed
-
- Reinhart B. J. et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906 (2000). - PubMed
-
- Tay Y., Zhang J., Thomson A. M., Lim B. & Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455, 1124–1128 (2008). - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

