PIK3CA mutations in androgen receptor-positive triple negative breast cancer confer sensitivity to the combination of PI3K and androgen receptor inhibitors
- PMID: 25103565
- PMCID: PMC4187324
- DOI: 10.1186/s13058-014-0406-x
PIK3CA mutations in androgen receptor-positive triple negative breast cancer confer sensitivity to the combination of PI3K and androgen receptor inhibitors
Abstract
Introduction: Triple negative breast cancer (TNBC) is a heterogeneous collection of biologically diverse cancers, which contributes to variable clinical outcomes. Previously, we identified a TNBC subtype that has a luminal phenotype and expresses the androgen receptor (AR+). TNBC cells derived from these luminal AR + tumors have high frequency phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) mutations. The purpose of this study was to determine if targeting phosphoinositide 3-kinase (PI3K) alone or in combination with an AR antagonist is effective in AR + TNBC.
Methods: We determined the frequency of activating PIK3CA mutations in AR + and AR- TNBC clinical cases. Using AR + TNBC cell line and xenograft models we evaluated the effectiveness of PI3K inhibitors, used alone or in combination with an AR antagonist, on tumor cell growth and viability.
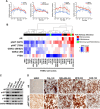
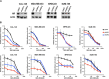
Results: PIK3CA kinase mutations were highly clonal, more frequent in AR + vs. AR- TNBC (40% vs. 4%), and often associated with concurrent amplification of the PIK3CA locus. PI3K/mTOR inhibitors had an additive growth inhibitory effect when combined with genetic or pharmacological AR targeting in AR + TNBC cells. We also analyzed the combination of bicalutamide +/- the pan-PI3K inhibitor GDC-0941 or the dual PI3K/mTOR inhibitor GDC-0980 in xenograft tumor studies and observed additive effects.
Conclusions: While approximately one third of TNBC patients respond to neoadjuvant/adjuvant chemotherapy, recent studies have shown that patients with AR + TNBC are far less likely to benefit from the current standard of care chemotherapy regimens and novel targeted approaches need to be investigated. In this study, we show that activating PIK3CA mutations are enriched in AR + TNBC; and, we show that the growth and viability of AR + TNBC cell line models is significantly reduced after treatment with PI3K inhibitors used in combination with an AR antagonist. These results provide rationale for pre-selection of TNBC patients with a biomarker (AR expression) to investigate the use of AR antagonists in combination with PI3K/mTOR inhibitors.
Figures




References
-
- Lin NU, Vanderplas A, Hughes ME, Theriault RL, Edge SB, Wong YN, Blayney DW, Niland JC, Winer EP, Weeks JC. Clinicopathologic features, patterns of recurrence, and survival among women with triple-negative breast cancer in the National Comprehensive Cancer Network. Cancer. 2012;118:5463–5472. doi: 10.1002/cncr.27581. - DOI - PMC - PubMed
-
- Keam B, Im SA, Kim HJ, Oh DY, Kim JH, Lee SH, Chie EK, Han W, Kim DW, Moon WK, Kim TY, Park IA, Noh DY, Heo DS, Ha SW, Bang YJ: Prognostic impact of clinicopathologic parameters in stage II/III breast cancer treated with neoadjuvant docetaxel and doxorubicin chemotherapy: paradoxical features of the triple negative breast cancer.BMC Cancer 2007, 7:203., - PMC - PubMed
-
- Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, Bashashati A, Prentice LM, Khattra J, Burleigh A, Yap D, Bernard V, McPherson A, Shumansky K, Crisan A, Giuliany R, Heravi-Moussavi A, Rosner J, Lai D, Birol I, Varhol R, Tam A, Dhalla N, Zeng T, Ma K, Chan SK, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

