Understanding the genetic and epigenetic architecture in complex network of rice flowering pathways
- PMID: 25103896
- PMCID: PMC4259885
- DOI: 10.1007/s13238-014-0068-6
Understanding the genetic and epigenetic architecture in complex network of rice flowering pathways
Abstract
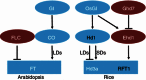
Although the molecular basis of flowering time control is well dissected in the long day (LD) plant Arabidopsis, it is still largely unknown in the short day (SD) plant rice. Rice flowering time (heading date) is an important agronomic trait for season adaption and grain yield, which is affected by both genetic and environmental factors. During the last decade, as the nature of florigen was identified, notable progress has been made on exploration how florigen gene expression is genetically controlled. In Arabidopsis expression of certain key flowering integrators such as FLOWERING LOCUS C (FLC) and FLOWERING LOCUS T (FT) are also epigenetically regulated by various chromatin modifications, however, very little is known in rice on this aspect until very recently. This review summarized the advances of both genetic networks and chromatin modifications in rice flowering time control, attempting to give a complete view of the genetic and epigenetic architecture in complex network of rice flowering pathways.
Figures
Similar articles
-
Enabling photoperiodic control of flowering by timely chromatin silencing of the florigen gene.Nucleus. 2015;6(3):179-82. doi: 10.1080/19491034.2015.1038000. Epub 2015 May 7. Nucleus. 2015. PMID: 25950625 Free PMC article.
-
Flowering time control in rice by introducing Arabidopsis clock-associated PSEUDO-RESPONSE REGULATOR 5.Biosci Biotechnol Biochem. 2020 May;84(5):970-979. doi: 10.1080/09168451.2020.1719822. Epub 2020 Jan 27. Biosci Biotechnol Biochem. 2020. PMID: 31985350
-
Chromatin regulation of flowering.Trends Plant Sci. 2012 Sep;17(9):556-62. doi: 10.1016/j.tplants.2012.05.001. Epub 2012 Jun 2. Trends Plant Sci. 2012. PMID: 22658650 Review.
-
A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice.Development. 2009 Oct;136(20):3443-50. doi: 10.1242/dev.040170. Epub 2009 Sep 17. Development. 2009. PMID: 19762423
-
Post-translational modifications of FLOWERING LOCUS C modulate its activity.J Exp Bot. 2017 Jan 1;68(3):383-389. doi: 10.1093/jxb/erw431. J Exp Bot. 2017. PMID: 28204510 Review.
Cited by
-
MORF-RELATED GENE702, a Reader Protein of Trimethylated Histone H3 Lysine 4 and Histone H3 Lysine 36, Is Involved in Brassinosteroid-Regulated Growth and Flowering Time Control in Rice.Plant Physiol. 2015 Aug;168(4):1275-85. doi: 10.1104/pp.114.255737. Epub 2015 Apr 8. Plant Physiol. 2015. PMID: 25855537 Free PMC article.
-
OsFTL4, an FT-like Gene, Regulates Flowering Time and Drought Tolerance in Rice (Oryza sativa L.).Rice (N Y). 2022 Sep 6;15(1):47. doi: 10.1186/s12284-022-00593-1. Rice (N Y). 2022. PMID: 36068333 Free PMC article.
-
Aphids and Mycorrhizal Fungi Shape Maternal Effects in Senecio vulgaris.Plants (Basel). 2022 Aug 18;11(16):2150. doi: 10.3390/plants11162150. Plants (Basel). 2022. PMID: 36015453 Free PMC article.
-
Hybrid Rice Production: A Worldwide Review of Floral Traits and Breeding Technology, with Special Emphasis on China.Plants (Basel). 2024 Feb 21;13(5):578. doi: 10.3390/plants13050578. Plants (Basel). 2024. PMID: 38475425 Free PMC article. Review.
-
The phosphoinositide-specific phospholipase C1 modulates flowering time and grain size in rice.Planta. 2022 Jul 4;256(2):29. doi: 10.1007/s00425-022-03941-z. Planta. 2022. PMID: 35781561
References
-
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, Goodman MM, Harjes C, Guill K, Kroon DE, Larsson S, Lepak NK, Li H, Mitchell SE, Pressoir G, Peiffer JA, Rosas MO, Rocheford TR, Romay MC, Romero S, Salvo S, Sanchez Villeda H, da Silva HS, Sun Q, Tian F, Upadyayula N, Ware D, Yates H, Yu J, Zhang Z, Kresovich S, McMullen MD. The genetic architecture of maize flowering time. Science. 2009;325:714–718. doi: 10.1126/science.1174276. - DOI - PubMed
-
- Cajlachjan MC. Concerning the hormonal nature of plant development processes. Compt Rend Acad Sci URSS. 1937;16:227–230.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

