Identifying causal variants at loci with multiple signals of association
- PMID: 25104515
- PMCID: PMC4196608
- DOI: 10.1534/genetics.114.167908
Identifying causal variants at loci with multiple signals of association
Abstract
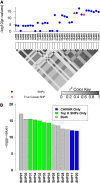
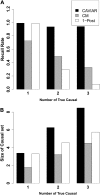
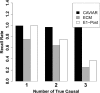
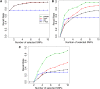
Although genome-wide association studies have successfully identified thousands of risk loci for complex traits, only a handful of the biologically causal variants, responsible for association at these loci, have been successfully identified. Current statistical methods for identifying causal variants at risk loci either use the strength of the association signal in an iterative conditioning framework or estimate probabilities for variants to be causal. A main drawback of existing methods is that they rely on the simplifying assumption of a single causal variant at each risk locus, which is typically invalid at many risk loci. In this work, we propose a new statistical framework that allows for the possibility of an arbitrary number of causal variants when estimating the posterior probability of a variant being causal. A direct benefit of our approach is that we predict a set of variants for each locus that under reasonable assumptions will contain all of the true causal variants with a high confidence level (e.g., 95%) even when the locus contains multiple causal variants. We use simulations to show that our approach provides 20-50% improvement in our ability to identify the causal variants compared to the existing methods at loci harboring multiple causal variants. We validate our approach using empirical data from an expression QTL study of CHI3L2 to identify new causal variants that affect gene expression at this locus. CAVIAR is publicly available online at http://genetics.cs.ucla.edu/caviar/.
Keywords: association studies; causal variants; fine mapping.
Copyright © 2014 by the Genetics Society of America.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- R01-GM083198/GM/NIGMS NIH HHS/United States
- R01 MH101782/MH/NIMH NIH HHS/United States
- U01-DA024417/DA/NIDA NIH HHS/United States
- P01 HL028481/HL/NHLBI NIH HHS/United States
- U01 DA024417/DA/NIDA NIH HHS/United States
- R03 CA162200/CA/NCI NIH HHS/United States
- R01-MH101782/MH/NIMH NIH HHS/United States
- K25-HL080079/HL/NHLBI NIH HHS/United States
- R01 ES022282/ES/NIEHS NIH HHS/United States
- K25 HL080079/HL/NHLBI NIH HHS/United States
- P30 NS062691/NS/NINDS NIH HHS/United States
- R01-ES022282/ES/NIEHS NIH HHS/United States
- P01-HL28481/HL/NHLBI NIH HHS/United States
- P01 HL030568/HL/NHLBI NIH HHS/United States
- R01 GM053275/GM/NIGMS NIH HHS/United States
- P01-HL30568/HL/NHLBI NIH HHS/United States
- R01 GM083198/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

