Comparison of pre-analytical FFPE sample preparation methods and their impact on massively parallel sequencing in routine diagnostics
- PMID: 25105902
- PMCID: PMC4126727
- DOI: 10.1371/journal.pone.0104566
Comparison of pre-analytical FFPE sample preparation methods and their impact on massively parallel sequencing in routine diagnostics
Abstract
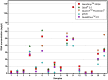
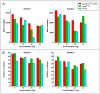
Over the last years, massively parallel sequencing has rapidly evolved and has now transitioned into molecular pathology routine laboratories. It is an attractive platform for analysing multiple genes at the same time with very little input material. Therefore, the need for high quality DNA obtained from automated DNA extraction systems has increased, especially to those laboratories which are dealing with formalin-fixed paraffin-embedded (FFPE) material and high sample throughput. This study evaluated five automated FFPE DNA extraction systems as well as five DNA quantification systems using the three most common techniques, UV spectrophotometry, fluorescent dye-based quantification and quantitative PCR, on 26 FFPE tissue samples. Additionally, the effects on downstream applications were analysed to find the most suitable pre-analytical methods for massively parallel sequencing in routine diagnostics. The results revealed that the Maxwell 16 from Promega (Mannheim, Germany) seems to be the superior system for DNA extraction from FFPE material. The extracts had a 1.3-24.6-fold higher DNA concentration in comparison to the other extraction systems, a higher quality and were most suitable for downstream applications. The comparison of the five quantification methods showed intermethod variations but all methods could be used to estimate the right amount for PCR amplification and for massively parallel sequencing. Interestingly, the best results in massively parallel sequencing were obtained with a DNA input of 15 ng determined by the NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). No difference could be detected in mutation analysis based on the results of the quantification methods. These findings emphasise, that it is particularly important to choose the most reliable and constant DNA extraction system, especially when using small biopsies and low elution volumes, and that all common DNA quantification techniques can be used for downstream applications like massively parallel sequencing.
Conflict of interest statement
Figures



References
-
- De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, et al. (2010) Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 11: 753–762. - PubMed
-
- Tuononen K, Mäki-Nevala S, Sarhadi VK, Wirtanen A, Rönty M, et al. (2013) Comparison of Targeted Next-Generation Sequencing (NGS) and Real-Time PCR in the Detection of EGFR, KRAS, and BRAF Mutations on Formalin-Fixed, Paraffin-Embedded Tumor Material of Non-Small Cell Lung Carcinoma—Superiority of NGS. Genes Chromosom Cancer 52: 503–511. - PubMed
-
- Hadd AG, Houghton J, Choudhary A, Sah S, Chen L, et al. (2013) Targeted, High-Depth, Next-Generation Sequencing of Cancer Genes in Formalin-Fixed, Paraffin-Embedded and Fine-Needle Aspiration Tumor Specimens. J Mol Diagn 15: 234–247. - PubMed
-
- Corless CL, Spellman PT (2012) Tackling Formalin-Fixed, Paraffin-Embedded Tumor Tissue with Next-Generation Sequencing. Cancer Discov 2: 23–24. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

