Correlation of phenotypic profiles using targeted proteomics identifies mycobacterial esx-1 substrates
- PMID: 25106450
- PMCID: PMC4227905
- DOI: 10.1021/pr500484w
Correlation of phenotypic profiles using targeted proteomics identifies mycobacterial esx-1 substrates
Abstract
The Esx/WXG-100 (ESAT-6/Wss) exporters are multiprotein complexes that promote protein translocation across the cytoplasmic membrane in a diverse range of pathogenic and nonpathogenic bacterial species. The Esx-1 (ESAT-6 System-1) system mediates virulence factor translocation in mycobacterial pathogens, including the human pathogen Mycobacterium tuberculosis. Although several genes have been associated with Esx-1-mediated transport and virulence, the contribution of individual Esx-1 genes to export is largely undefined. A unique aspect of Esx-1 export is that several substrates require each other for export/stability. We exploited substrate "codependency" to identify Esx-1 substrates. We simultaneously quantified changes in the levels of 13 Esx-1 proteins from both secreted and cytosolic protein fractions generated from 16 Esx-1-deficient Mycobacterium marinum strains in a single experiment using MRM/SRM targeted mass spectrometry. This expansion of measurable Esx-1 proteins allowed us to define statistical rules for assigning novel substrates using phenotypic profiles of known Esx-1 substrates. Using this approach, we identified three additional Esx-1 substrates encoded by the esx-1 region. Our studies begin to address how disruption of specific genes affects several proteins in the Esx-1 complex. Overall, our findings illuminate relationships between Esx-1 proteins and create a framework for the identification of secreted substrates applicable to other protein exporters and pathways.
Keywords: Esx-1; EsxA; MRM/SRM; Mycobacterium marinum; RD1; secretion; substrate identification; targeted proteomics.
Figures



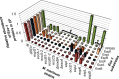
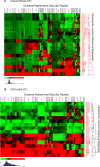
References
-
- BCG Vaccine; World Health Organization: Geneva, 2009.
-
- Tuberculosis & Diabetes: Collaborative framework for care and control of tuberculosis and diabetes; World Health Organization: Geneva, 2011. - PubMed
-
- WHO Global Tuberculosis Report 2013; World Health Organization: Geneva, 2013.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

