The CHR site: definition and genome-wide identification of a cell cycle transcriptional element
- PMID: 25106871
- PMCID: PMC4176359
- DOI: 10.1093/nar/gku696
The CHR site: definition and genome-wide identification of a cell cycle transcriptional element
Abstract
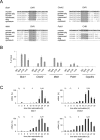
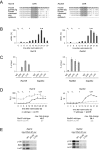
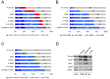
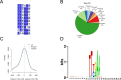
The cell cycle genes homology region (CHR) has been identified as a DNA element with an important role in transcriptional regulation of late cell cycle genes. It has been shown that such genes are controlled by DREAM, MMB and FOXM1-MuvB and that these protein complexes can contact DNA via CHR sites. However, it has not been elucidated which sequence variations of the canonical CHR are functional and how frequent CHR-based regulation is utilized in mammalian genomes. Here, we define the spectrum of functional CHR elements. As the basis for a computational meta-analysis, we identify new CHR sequences and compile phylogenetic motif conservation as well as genome-wide protein-DNA binding and gene expression data. We identify CHR elements in most late cell cycle genes binding DREAM, MMB, or FOXM1-MuvB. In contrast, Myb- and forkhead-binding sites are underrepresented in both early and late cell cycle genes. Our findings support a general mechanism: sequential binding of DREAM, MMB and FOXM1-MuvB complexes to late cell cycle genes requires CHR elements. Taken together, we define the group of CHR-regulated genes in mammalian genomes and provide evidence that the CHR is the central promoter element in transcriptional regulation of late cell cycle genes by DREAM, MMB and FOXM1-MuvB.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Müller G.A., Engeland K. The central role of CDE/CHR promoter elements in the regulation of cell cycle-dependent gene transcription. FEBS J. 2010;277:877–893. - PubMed
-
- Schmit F., Cremer S., Gaubatz S. LIN54 is an essential core subunit of the DREAM/LINC complex that binds to the cdc2 promoter in a sequence-specific manner. FEBS J. 2009;276:5703–5716. - PubMed
-
- Litovchick L., Sadasivam S., Florens L., Zhu X., Swanson S.K., Velmurugan S., Chen R., Washburn M.P., Liu X.S., DeCaprio J.A. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell. 2007;26:539–551. - PubMed
-
- Schmit F., Korenjak M., Mannefeld M., Schmitt K., Franke C., von E.B., Gagrica S., Hanel F., Brehm A., Gaubatz S. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle. 2007;6:1903–1913. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

