Upregulation of human PINK1 gene expression by NFκB signalling
- PMID: 25108683
- PMCID: PMC4237968
- DOI: 10.1186/s13041-014-0057-y
Upregulation of human PINK1 gene expression by NFκB signalling
Abstract
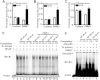
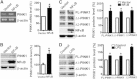
Parkinson's disease (PD) is one of the major neurodegenerative disorders. Mitochondrial malfunction is implicated in PD pathogenesis. Phosphatase and tensin homolog deleted on chromosome 10 (PTEN)-induced putative kinase 1 (PINK1), a serine/threonine kinase, plays an important role in the quality control of mitochondria and more than 70 PINK1 mutations have been identified to cause early-onset PD. However, the regulation of PINK1 gene expression remains elusive. In the present study, we identified the transcription start site (TSS) of the human PINK1 gene using switching mechanism at 5'end of RNA transcription (SMART RACE) assay. The TSS is located at 91 bp upstream of the translation start site ATG. The region with 104 bp was identified as the minimal promoter region by deletion analysis followed by dual luciferase assay. Four functional cis-acting nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB)-binding sites within the PINK1 promoter were identified. NFκB overexpression led to the up-regulation of PINK1 expression in both HEK293 cells and SH-SY5Y cells. Consistently, lipopolysaccharide (LPS), a strong activator of NFκB, significantly increased PINK1 expression in SH-SY5Y cells. Taken together, our results clearly suggested that PINK1 expression is tightly regulated at its transcription level and NFκB is a positive regulator for PINK1 expression.
Figures


References
-
- Parkinson J. An essay on the shaking palsy. 1817. J Neuropsychiatry Clin Neurosci. 2002;14(2):223–236. discussion 222. - PubMed
-
- Lewy F. Paralysis agitans. I. Pathologische anatomie. Handbuch der Neurologie. 1912;3:920–933.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

