Contribution of human immunodeficiency virus type 1 minority variants to reduced drug susceptibility in patients on an integrase strand transfer inhibitor-based therapy
- PMID: 25110880
- PMCID: PMC4128663
- DOI: 10.1371/journal.pone.0104512
Contribution of human immunodeficiency virus type 1 minority variants to reduced drug susceptibility in patients on an integrase strand transfer inhibitor-based therapy
Abstract
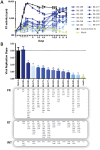
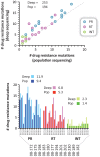
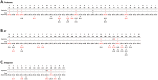
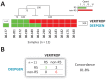
The role of HIV-1 minority variants on transmission, pathogenesis, and virologic failure to antiretroviral regimens has been explored; however, most studies of low-level HIV-1 drug-resistant variants have focused in single target regions. Here we used a novel HIV-1 genotypic assay based on deep sequencing, DEEPGEN (Gibson et al 2014 Antimicrob Agents Chemother 58∶2167) to simultaneously analyze the presence of minority variants carrying mutations associated with reduced susceptibility to protease (PR), reverse transcriptase (RT), and integrase strand transfer integrase inhibitors (INSTIs), as well as HIV-1 coreceptor tropism. gag-p2/NCp7/p1/p6/pol-PR/RT/INT and env/C2V3 PCR products were obtained from twelve heavily treatment-experienced patients experiencing virologic failure while participating in a 48-week dose-ranging study of elvitegravir (GS-US-183-0105). Deep sequencing results were compared with (i) virological response to treatment, (ii) genotyping based on population sequencing, (iii) phenotyping data using PhenoSense and VIRALARTS, and (iv) HIV-1 coreceptor tropism based on the phenotypic test VERITROP. Most patients failed the antiretroviral regimen with numerous pre-existing mutations in the PR and RT, and additionally newly acquired INSTI-resistance mutations as determined by population sequencing (mean 9.4, 5.3, and 1.4 PI- RTI-, and INSTI-resistance mutations, respectively). Interestingly, since DEEPGEN allows the accurate detection of amino acid substitutions at frequencies as low as 1% of the population, a series of additional drug resistance mutations were detected by deep sequencing (mean 2.5, 1.5, and 0.9, respectively). The presence of these low-abundance HIV-1 variants was associated with drug susceptibility, replicative fitness, and coreceptor tropism determined using sensitive phenotypic assays, enhancing the overall burden of resistance to all four antiretroviral drug classes. Further longitudinal studies based on deep sequencing tests will help to clarify (i) the potential impact of minority HIV-1 drug resistant variants in response to antiretroviral therapy and (ii) the importance of the detection of HIV minority variants in the clinical practice.
Conflict of interest statement
Figures


Similar articles
-
Sensitive deep-sequencing-based HIV-1 genotyping assay to simultaneously determine susceptibility to protease, reverse transcriptase, integrase, and maturation inhibitors, as well as HIV-1 coreceptor tropism.Antimicrob Agents Chemother. 2014;58(4):2167-85. doi: 10.1128/AAC.02710-13. Epub 2014 Jan 27. Antimicrob Agents Chemother. 2014. PMID: 24468782 Free PMC article.
-
Resistance mutations outside the integrase coding region have an effect on human immunodeficiency virus replicative fitness but do not affect its susceptibility to integrase strand transfer inhibitors.PLoS One. 2013 Jun 11;8(6):e65631. doi: 10.1371/journal.pone.0065631. Print 2013. PLoS One. 2013. PMID: 23776513 Free PMC article.
-
Altered viral fitness and drug susceptibility in HIV-1 carrying mutations that confer resistance to nonnucleoside reverse transcriptase and integrase strand transfer inhibitors.J Virol. 2014 Aug;88(16):9268-76. doi: 10.1128/JVI.00695-14. Epub 2014 Jun 4. J Virol. 2014. PMID: 24899199 Free PMC article.
-
Study of the impact of HIV genotypic drug resistance testing on therapy efficacy.Verh K Acad Geneeskd Belg. 2001;63(5):447-73. Verh K Acad Geneeskd Belg. 2001. PMID: 11813503 Review.
-
HIV drug resistance against strand transfer integrase inhibitors.Retrovirology. 2017 Jun 5;14(1):36. doi: 10.1186/s12977-017-0360-7. Retrovirology. 2017. PMID: 28583191 Free PMC article. Review.
Cited by
-
Nanoscale flow cytometry reveals interpatient variability in HIV protease activity that correlates with viral infectivity and identifies drug-resistant viruses.Sci Rep. 2020 Oct 22;10(1):18101. doi: 10.1038/s41598-020-75118-1. Sci Rep. 2020. PMID: 33093566 Free PMC article.
-
Characterization of minority HIV-1 drug resistant variants in the United Kingdom following the verification of a deep sequencing-based HIV-1 genotyping and tropism assay.AIDS Res Ther. 2018 Nov 8;15(1):18. doi: 10.1186/s12981-018-0206-y. AIDS Res Ther. 2018. PMID: 30409215 Free PMC article.
-
Low-Frequency Drug Resistance in HIV-Infected Ugandans on Antiretroviral Treatment Is Associated with Regimen Failure.Antimicrob Agents Chemother. 2016 May 23;60(6):3380-97. doi: 10.1128/AAC.00038-16. Print 2016 Jun. Antimicrob Agents Chemother. 2016. PMID: 27001818 Free PMC article.
-
A bibliometric analysis of HIV-1 drug-resistant minority variants from 1999 to 2024.AIDS Res Ther. 2025 Apr 10;22(1):47. doi: 10.1186/s12981-025-00739-3. AIDS Res Ther. 2025. PMID: 40211381 Free PMC article.
-
Impaired human immunodeficiency virus type 1 replicative fitness in atypical viremic non-progressor individuals.AIDS Res Ther. 2017 Mar 20;14:15. doi: 10.1186/s12981-017-0144-0. eCollection 2017. AIDS Res Ther. 2017. PMID: 28331526 Free PMC article.
References
-
- Paredes R, Clotet B (2010) Clinical management of HIV-1 resistance. Antiviral Res 85: 245–265. - PubMed
-
- Zolopa AR (2010) The evolution of HIV treatment guidelines: current state-of-the-art of ART. Antiviral Res 85: 241–244. - PubMed
-
- Fast PE, Price MA, Rida WN, Kamali A, Karita E (2013) WHO's new guidelines for antiretroviral treatment. Lancet 382: 1778–1779. - PubMed
-
- Summa V, Petrocchi A, Bonelli F, Crescenzi B, Donghi M, et al. (2008) Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. Journal of medicinal chemistry 51: 5843–5855. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

