Population genomics of the fission yeast Schizosaccharomyces pombe
- PMID: 25111393
- PMCID: PMC4128662
- DOI: 10.1371/journal.pone.0104241
Population genomics of the fission yeast Schizosaccharomyces pombe
Abstract
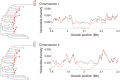
The fission yeast Schizosaccharomyces pombe has been widely used as a model eukaryote to study a diverse range of biological processes. However, population genetic studies of this species have been limited to date, and we know very little about the evolutionary processes and selective pressures that are shaping its genome. Here, we sequenced the genomes of 32 worldwide S. pombe strains and examined the pattern of polymorphisms across their genomes. In addition to introns and untranslated regions (UTRs), intergenic regions also exhibited lower levels of nucleotide diversity than synonymous sites, suggesting that a considerable amount of noncoding DNA is under selective constraint and thus likely to be functional. A number of genomic regions showed a reduction of nucleotide diversity probably caused by selective sweeps. We also identified a region close to the end of chromosome 3 where an extremely high level of divergence was observed between 5 of the 32 strains and the remain 27, possibly due to introgression, strong positive selection, or that region being responsible for reproductive isolation. Our study should serve as an important starting point in using a population genomics approach to further elucidate the biology of this important model organism.
Conflict of interest statement
Figures
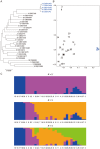
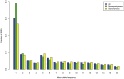
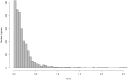
 ratios of
ratios of  are also not shown.
are also not shown.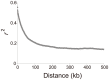
 is plotted as a function of distance in kilobase up to 500 kb. The average
is plotted as a function of distance in kilobase up to 500 kb. The average  values over each non-overlapping 1 kb bin are shown.
values over each non-overlapping 1 kb bin are shown.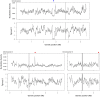

References
-
- Nurse P (1990) Universal control mechanism regulating onset of M-phase. Nature 344: 503–508. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

