Guiding functional connectivity estimation by structural connectivity in MEG: an application to discrimination of conditions of mild cognitive impairment
- PMID: 25111472
- PMCID: PMC4312351
- DOI: 10.1016/j.neuroimage.2014.08.002
Guiding functional connectivity estimation by structural connectivity in MEG: an application to discrimination of conditions of mild cognitive impairment
Abstract
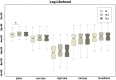
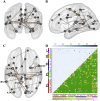
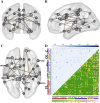
Whole brain resting state connectivity is a promising biomarker that might help to obtain an early diagnosis in many neurological diseases, such as dementia. Inferring resting-state connectivity is often based on correlations, which are sensitive to indirect connections, leading to an inaccurate representation of the real backbone of the network. The precision matrix is a better representation for whole brain connectivity, as it considers only direct connections. The network structure can be estimated using the graphical lasso (GL), which achieves sparsity through l1-regularization on the precision matrix. In this paper, we propose a structural connectivity adaptive version of the GL, where weaker anatomical connections are represented as stronger penalties on the corresponding functional connections. We applied beamformer source reconstruction to the resting state MEG recordings of 81 subjects, where 29 were healthy controls, 22 were single-domain amnestic Mild Cognitive Impaired (MCI), and 30 were multiple-domain amnestic MCI. An atlas-based anatomical parcellation of 66 regions was obtained for each subject, and time series were assigned to each of the regions. The fiber densities between the regions, obtained with deterministic tractography from diffusion-weighted MRI, were used to define the anatomical connectivity. Precision matrices were obtained with the region specific time series in five different frequency bands. We compared our method with the traditional GL and a functional adaptive version of the GL, in terms of log-likelihood and classification accuracies between the three groups. We conclude that introducing an anatomical prior improves the expressivity of the model and, in most cases, leads to a better classification between groups.
Keywords: Diffusion tensor imaging; Graphical Lasso; Machine learning; Magnetoencephalography; Mild cognitive impairment; Multimodal neuroimaging; Multivariate sparse regression; Resting state.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures



References
-
- Auer S., Reisberg B. The GDS/FAST staging system. Int. Psychogeriatr. 1997;9(Suppl. 1):167–171. - PubMed
-
- Bajo R., Maestú F., Nevado A., Sancho M., Gutiérrez R., Campo P., Castellanos N.P., Gil P., Moratti S., Pereda E., Del-Pozo F. Functional connectivity in mild cognitive impairment during a memory task: implications for the disconnection hypothesis. J. Alzheimers Dis. 2010;22:183–193. - PubMed
-
- Bajo R., Castellanos N.P., Cuesta P., Aurtenetxe S., Garcia-Prieto J., Gil-Gregorio P., del-Pozo F., Maestu F. Differential patterns of connectivity in progressive mild cognitive impairment. Brain Connect. 2012;2:21–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

