A novel high-resolution single locus sequence typing scheme for mixed populations of Propionibacterium acnes in vivo
- PMID: 25111794
- PMCID: PMC4128656
- DOI: 10.1371/journal.pone.0104199
A novel high-resolution single locus sequence typing scheme for mixed populations of Propionibacterium acnes in vivo
Abstract
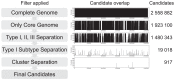
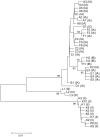
The Gram-positive anaerobic bacterium Propionibacterium acnes is a prevalent member of the normal skin microbiota of human adults. In addition to its suspected role in acne vulgaris it is involved in a variety of opportunistic infections. Multi-locus sequence-typing (MLST) schemes identified distinct phylotypes associated with health and disease. Being based on 8 to 9 house-keeping genes these MLST schemes have a high discriminatory power, but their application is time- and cost-intensive. Here we describe a single-locus sequence typing (SLST) scheme for P. acnes. The target locus was identified with a genome mining approach that took advantage of the availability of representative genome sequences of all known phylotypes of P. acnes. We applied this SLST on a collection of 188 P. acnes strains and demonstrated a resolution comparable to that of existing MLST schemes. Phylogenetic analysis applied to the SLST locus resulted in clustering patterns identical to a reference tree based on core genome sequences. We further demonstrate that SLST can be applied to detect multiple phylotypes in complex microbial communities by a metagenomic pyrosequencing approach. The described SLST strategy may be applied to any bacterial species with a basically clonal population structure to achieve easy typing and mapping of multiple phylotypes in complex microbiotas. The P. acnes SLST database can be found at http://medbac.dk/slst/pacnes.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

