Competence for genetic transformation in Streptococcus pneumoniae: mutations in σA bypass the comW requirement
- PMID: 25112479
- PMCID: PMC4248798
- DOI: 10.1128/JB.01933-14
Competence for genetic transformation in Streptococcus pneumoniae: mutations in σA bypass the comW requirement
Abstract
Competence for genetic transformation in the genus Streptococcus depends on an alternative sigma factor, σ(X), for coordinated synthesis of 23 proteins, which together establish the X state by permitting lysis of incompetent streptococci, uptake of DNA fragments, and integration of strands of that DNA into the resident genome. Initiation of transient accumulation of high levels of σ(X) is coordinated between cells by transcription factors linked to peptide pheromone signals. In Streptococcus pneumoniae, elevated σ(X) is insufficient for development of full competence without coexpression of a second competence-specific protein, ComW. ComW, shared by eight species in the Streptococcus mitis and Streptococcus anginosus groups, is regulated by the same pheromone circuit that controls σ(X), but its role in expression of the σ(X) regulon is unknown. Using the strong, but not absolute, dependence of transformation on comW as a selective tool, we collected 27 independent comW bypass mutations and mapped them to 10 single-base transitions, all within rpoD, encoding the primary sigma factor subunit of RNA polymerase, σ(A). Eight mapped to sites in rpoD region 4 that are implicated in interaction with the core β subunit, indicating that ComW may act to facilitate competition of the alternative sigma factor σ(X) for access to core polymerase.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures
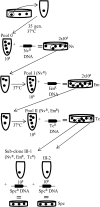
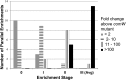



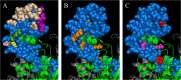
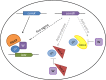
References
-
- Lacks SA. 2004. Transformation, p 89–115 In Mitchell TJ, Morrison DA, Spratt BG, Tuomanen EI. (ed), The pneumococcus. ASM Press, Washington, DC.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

