Enhanced killing of antibiotic-resistant bacteria enabled by massively parallel combinatorial genetics
- PMID: 25114216
- PMCID: PMC4151723
- DOI: 10.1073/pnas.1400093111
Enhanced killing of antibiotic-resistant bacteria enabled by massively parallel combinatorial genetics
Abstract
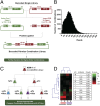
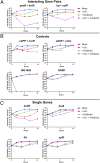
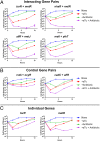
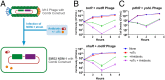
New therapeutic strategies are needed to treat infections caused by drug-resistant bacteria, which constitute a major growing threat to human health. Here, we use a high-throughput technology to identify combinatorial genetic perturbations that can enhance the killing of drug-resistant bacteria with antibiotic treatment. This strategy, Combinatorial Genetics En Masse (CombiGEM), enables the rapid generation of high-order barcoded combinations of genetic elements for high-throughput multiplexed characterization based on next-generation sequencing. We created ∼ 34,000 pairwise combinations of Escherichia coli transcription factor (TF) overexpression constructs. Using Illumina sequencing, we identified diverse perturbations in antibiotic-resistance phenotypes against carbapenem-resistant Enterobacteriaceae. Specifically, we found multiple TF combinations that potentiated antibiotic killing by up to 10(6)-fold and delivered these combinations via phagemids to increase the killing of highly drug-resistant E. coli harboring New Delhi metallo-beta-lactamase-1. Moreover, we constructed libraries of three-wise combinations of transcription factors with >4 million unique members and demonstrated that these could be tracked via next-generation sequencing. We envision that CombiGEM could be extended to other model organisms, disease models, and phenotypes, where it could accelerate massively parallel combinatorial genetics studies for a broad range of biomedical and biotechnology applications, including the treatment of antibiotic-resistant infections.
Keywords: combination therapy; drug resistance; multifactorial genetics; synthetic biology; systems biology.
Conflict of interest statement
Conflict of interest statement: The authors have submitted a patent application for this technology.
Figures
Similar articles
-
Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM.Proc Natl Acad Sci U S A. 2016 Mar 1;113(9):2544-9. doi: 10.1073/pnas.1517883113. Epub 2016 Feb 10. Proc Natl Acad Sci U S A. 2016. PMID: 26864203 Free PMC article.
-
Polymyxin Combinations Combat Escherichia coli Harboring mcr-1 and blaNDM-5: Preparation for a Postantibiotic Era.mBio. 2017 Jul 25;8(4):e00540-17. doi: 10.1128/mBio.00540-17. mBio. 2017. PMID: 28743810 Free PMC article.
-
Detection of Antibiotic Resistance Determinants and Their Transmissibility among Clinically Isolated Carbapenem-Resistant Escherichia coli from South India.Med Princ Pract. 2018;27(5):428-435. doi: 10.1159/000489885. Epub 2018 May 8. Med Princ Pract. 2018. PMID: 29739004 Free PMC article.
-
The threat of carbapenem-resistant bacteria in the environment: Evidence of widespread contamination of reservoirs at a global scale.Environ Pollut. 2019 Dec;255(Pt 1):113143. doi: 10.1016/j.envpol.2019.113143. Epub 2019 Sep 9. Environ Pollut. 2019. PMID: 31541827 Review.
-
Current epidemiology, genetic evolution and clinical impact of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae.Infect Genet Evol. 2018 Jul;61:185-188. doi: 10.1016/j.meegid.2018.04.005. Epub 2018 Apr 5. Infect Genet Evol. 2018. PMID: 29626676 Review.
Cited by
-
Mechanism of Action of Isopropoxy Benzene Guanidine against Multidrug-Resistant Pathogens.Microbiol Spectr. 2023 Feb 14;11(1):e0346922. doi: 10.1128/spectrum.03469-22. Epub 2022 Dec 8. Microbiol Spectr. 2023. PMID: 36475769 Free PMC article.
-
Corynebacterium glutamicum Metabolic Engineering with CRISPR Interference (CRISPRi).ACS Synth Biol. 2016 May 20;5(5):375-85. doi: 10.1021/acssynbio.5b00216. Epub 2016 Feb 16. ACS Synth Biol. 2016. PMID: 26829286 Free PMC article.
-
Tackling Drug Resistant Infection Outbreaks of Global Pandemic Escherichia coli ST131 Using Evolutionary and Epidemiological Genomics.Microorganisms. 2015 May 20;3(2):236-67. doi: 10.3390/microorganisms3020236. Microorganisms. 2015. PMID: 27682088 Free PMC article. Review.
-
Engineering allostery.Trends Genet. 2014 Dec;30(12):521-8. doi: 10.1016/j.tig.2014.09.004. Epub 2014 Oct 8. Trends Genet. 2014. PMID: 25306102 Free PMC article. Review.
-
A Barcoding Strategy Enabling Higher-Throughput Library Screening by Microscopy.ACS Synth Biol. 2015 Nov 20;4(11):1205-16. doi: 10.1021/acssynbio.5b00060. Epub 2015 Jul 15. ACS Synth Biol. 2015. PMID: 26155738 Free PMC article.
References
-
- Centers for Disease Control and Prevention 2013. Antibiotic resistance threats in the US. Available at www.cdc.gov/drugresistance/threat-report-2013/. Accessed July 21, 2014.
-
- Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6(1):29–40. - PubMed
-
- Leverstein-Van Hall MA, et al. Global spread of New Delhi metallo-β-lactamase 1. Lancet Infect Dis. 2010;10(12):830–831. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

