A new tubulin-binding site and pharmacophore for microtubule-destabilizing anticancer drugs
- PMID: 25114240
- PMCID: PMC4183314
- DOI: 10.1073/pnas.1408124111
A new tubulin-binding site and pharmacophore for microtubule-destabilizing anticancer drugs
Abstract
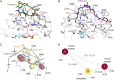
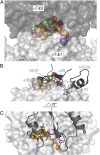
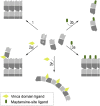
The recent success of antibody-drug conjugates (ADCs) in the treatment of cancer has led to a revived interest in microtubule-destabilizing agents. Here, we determined the high-resolution crystal structure of the complex between tubulin and maytansine, which is part of an ADC that is approved by the US Food and Drug Administration (FDA) for the treatment of advanced breast cancer. We found that the drug binds to a site on β-tubulin that is distinct from the vinca domain and that blocks the formation of longitudinal tubulin interactions in microtubules. We also solved crystal structures of tubulin in complex with both a variant of rhizoxin and the phase 1 drug PM060184. Consistent with biochemical and mutagenesis data, we found that the two compounds bound to the same site as maytansine and that the structures revealed a common pharmacophore for the three ligands. Our results delineate a distinct molecular mechanism of action for the inhibition of microtubule assembly by clinically relevant agents. They further provide a structural basis for the rational design of potent microtubule-destabilizing agents, thus opening opportunities for the development of next-generation ADCs for the treatment of cancer.
Keywords: X-ray crystallography; drug mechanism; microtubule-targeting agents.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
A previously undescribed tubulin binder.Proc Natl Acad Sci U S A. 2014 Sep 23;111(38):13684-5. doi: 10.1073/pnas.1414572111. Epub 2014 Sep 3. Proc Natl Acad Sci U S A. 2014. PMID: 25187564 Free PMC article. No abstract available.
References
-
- Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004;4(4):253–265. - PubMed
-
- Canta A, Chiorazzi A, Cavaletti G. Tubulin: A target for antineoplastic drugs into the cancer cells but also in the peripheral nervous system. Curr Med Chem. 2009;16(11):1315–1324. - PubMed
-
- Zolot RS, Basu S, Million RP. Antibody-drug conjugates. Nat Rev Drug Discov. 2013;12(4):259–260. - PubMed
-
- Teicher BA, Doroshow JH. The promise of antibody-drug conjugates. N Engl J Med. 2012;367(19):1847–1848. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

