Genetic network identifies novel pathways contributing to atherosclerosis susceptibility in the innominate artery
- PMID: 25115202
- PMCID: PMC4142055
- DOI: 10.1186/1755-8794-7-51
Genetic network identifies novel pathways contributing to atherosclerosis susceptibility in the innominate artery
Abstract
Background: Atherosclerosis, the underlying cause of cardiovascular disease, results from both genetic and environmental factors.
Methods: In the current study we take a systems-based approach using weighted gene co-expression analysis to identify a candidate pathway of genes related to atherosclerosis. Bioinformatic analyses are performed to identify candidate genes and interactions and several novel genes are characterized using in-vitro studies.
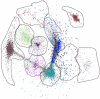
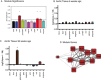
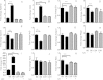
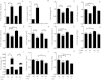
Results: We identify 1 coexpression module associated with innominate artery atherosclerosis that is also enriched for inflammatory and macrophage gene signatures. Using a series of bioinformatics analysis, we further prioritize the genes in this pathway and identify Cd44 as a critical mediator of the atherosclerosis. We validate our predictions generated by the network analysis using Cd44 knockout mice.
Conclusion: These results indicate that alterations in Cd44 expression mediate inflammation through a complex transcriptional network involving a number of previously uncharacterized genes.
Figures


Similar articles
-
Effect of macrophage overexpression of murine liver X receptor-alpha (LXR-alpha) on atherosclerosis in LDL-receptor deficient mice.Arterioscler Thromb Vasc Biol. 2008 Nov;28(11):2009-15. doi: 10.1161/ATVBAHA.108.175257. Epub 2008 Sep 11. Arterioscler Thromb Vasc Biol. 2008. PMID: 18787185
-
Multi-organ expression profiling uncovers a gene module in coronary artery disease involving transendothelial migration of leukocytes and LIM domain binding 2: the Stockholm Atherosclerosis Gene Expression (STAGE) study.PLoS Genet. 2009 Dec;5(12):e1000754. doi: 10.1371/journal.pgen.1000754. Epub 2009 Dec 4. PLoS Genet. 2009. PMID: 19997623 Free PMC article.
-
Transcriptome analysis of genes regulated by cholesterol loading in two strains of mouse macrophages associates lysosome pathway and ER stress response with atherosclerosis susceptibility.PLoS One. 2013 May 21;8(5):e65003. doi: 10.1371/journal.pone.0065003. Print 2013. PLoS One. 2013. PMID: 23705026 Free PMC article.
-
Investigation of the underlying genes and mechanism of familial hypercholesterolemia through bioinformatics analysis.BMC Cardiovasc Disord. 2020 Sep 16;20(1):419. doi: 10.1186/s12872-020-01701-z. BMC Cardiovasc Disord. 2020. PMID: 32938406 Free PMC article.
-
Weighted gene co-expression network analysis identifies specific modules and hub genes related to coronary artery disease.BMC Cardiovasc Disord. 2016 Mar 5;16:54. doi: 10.1186/s12872-016-0217-3. BMC Cardiovasc Disord. 2016. PMID: 26944061 Free PMC article.
Cited by
-
Signature transcriptome analysis of stage specific atherosclerotic plaques of patients.BMC Med Genomics. 2022 Apr 29;15(1):99. doi: 10.1186/s12920-022-01250-8. BMC Med Genomics. 2022. PMID: 35488341 Free PMC article.
-
Analytical Strategy to Prioritize Alzheimer's Disease Candidate Genes in Gene Regulatory Networks Using Public Expression Data.J Alzheimers Dis. 2017;59(4):1237-1254. doi: 10.3233/JAD-170011. J Alzheimers Dis. 2017. PMID: 28800327 Free PMC article.
-
Macrophage Trafficking, Inflammatory Resolution, and Genomics in Atherosclerosis: JACC Macrophage in CVD Series (Part 2).J Am Coll Cardiol. 2018 Oct 30;72(18):2181-2197. doi: 10.1016/j.jacc.2018.08.2147. J Am Coll Cardiol. 2018. PMID: 30360827 Free PMC article. Review.
-
Obesogenic and diabetic effects of CD44 in mice are sexually dimorphic and dependent on genetic background.Biol Sex Differ. 2022 Apr 11;13(1):14. doi: 10.1186/s13293-022-00426-2. Biol Sex Differ. 2022. PMID: 35410390 Free PMC article.
-
Upregulation of TH/IL-17 Pathway-Related Genes in Human Coronary Endothelial Cells Stimulated with Serum of Patients with Acute Coronary Syndromes.Front Cardiovasc Med. 2017 Feb 7;4:1. doi: 10.3389/fcvm.2017.00001. eCollection 2017. Front Cardiovasc Med. 2017. PMID: 28224128 Free PMC article.
References
-
- Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Franco S, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Huffman M, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Magid D, Marcus GM, Marelli A, Matchar DB, McGuire DK, Mohler ER, Moy CS, Mussolino ME. et al.Executive summary: heart disease and stroke statistics–2013 update: a report from the American Heart Association. Circulation. 2013;7:143–152. - PubMed
-
- Lusis AJ, Mar R, Pajukanta P. Genetics of atherosclerosis. Annu Rev Genomics Hum Genet. 2004;7:189–218. - PubMed
-
- Homma S, Ishii T, Malcom GT, Zieske AW, Strong JP, Tsugane S, Hirose N. Histopathological modifications of early atherosclerotic lesions by risk factors–findings in PDAY subjects. Atherosclerosis. 2001;7:389–399. - PubMed
-
- Yan ZQ, Hansson GK. Innate immunity, macrophage activation, and atherosclerosis. Immunol Rev. 2007;7:187–203. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

