A 5-gene classifier from the carcinoma-associated fibroblast transcriptomic profile and clinical outcome in colorectal cancer
- PMID: 25115384
- PMCID: PMC4171642
- DOI: 10.18632/oncotarget.2237
A 5-gene classifier from the carcinoma-associated fibroblast transcriptomic profile and clinical outcome in colorectal cancer
Abstract
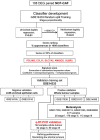
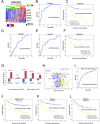
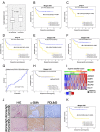
Based on 108 differentially expressed genes between carcinoma-associated fibroblasts (CAFs) and paired normal colonic fibroblasts we recently reported, a 5-gene classifier for relapse prediction in Stage II/III colorectal cancer (CRC ) was developed. Its predictive value was validated in datasets GSE17538, GSE33113 and GSE14095. An additional validation was performed in a metacohort (n=317) and 142 CRC patients by means of RT-PCR. The 5-gene classifier was significantly associated with increased relapse risk and death from CRC across all validation series of Stage II/III patients used. Multivariate Cox regression analyses confirmed the independent prognostic value of the stromal classifier (HR=2.67; P=0.002). Post-test probabilities provided evidence of the suitability of the 5-gene classifier in clinical practice, identifying a subgroup of Stage-II patients who were at high risk of relapse. Moreover, the a priory worst prognosis mesenchymal subtype of tumours can be stratified according to the physiological status of their carcinoma-associated fibroblasts. In conclusion the CAFs-derived 5-gene classifier provides more accurate information about outcome than conventional clinicopathological criteria and it could be useful to take clinical decisions, especially in Stage II. Additionally, the classifier put into relevance the CAF's intratumoral heterogeneity and might contribute to find relevant targets for depleting adequate CAFS subtypes.
Conflict of interest statement
The is no conflict of interest of any form.
Figures
References
-
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 127;12(2893):2917–2917. - PubMed
-
- O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96(19):1420–1420. - PubMed
-
- Andre T, Boni C, Mounedji-Boudiaf L, Navarro M, Tabernero J, Hickish T, Topham C, Zaninelli M, Clingan P, Bridgewater J, Tabah-Fisch I, de Gramont A. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. The New England journal of medicine. 2004;350(23):2343–2343. - PubMed
-
- Gray R, Barnwell J, McConkey C, Hills RK, Williams NS, Kerr DJ. Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet. 2007;370(9604):2020–2020. - PubMed
-
- Ragnhammar P, Hafstrom L, Nygren P, Glimelius B. A systematic overview of chemotherapy effects in colorectal cancer. Acta oncologica (Stockholm, Sweden) 2001;40(2-3):282–308. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

