Approaches to integrating germline and tumor genomic data in cancer research
- PMID: 25115441
- PMCID: PMC4178473
- DOI: 10.1093/carcin/bgu165
Approaches to integrating germline and tumor genomic data in cancer research
Abstract
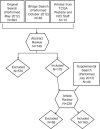
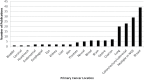
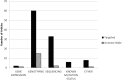
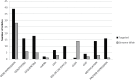
Cancer is characterized by a diversity of genetic and epigenetic alterations occurring in both the germline and somatic (tumor) genomes. Hundreds of germline variants associated with cancer risk have been identified, and large amounts of data identifying mutations in the tumor genome that participate in tumorigenesis have been generated. Increasingly, these two genomes are being explored jointly to better understand how cancer risk alleles contribute to carcinogenesis and whether they influence development of specific tumor types or mutation profiles. To understand how data from germline risk studies and tumor genome profiling is being integrated, we reviewed 160 articles describing research that incorporated data from both genomes, published between January 2009 and December 2012, and summarized the current state of the field. We identified three principle types of research questions being addressed using these data: (i) use of tumor data to determine the putative function of germline risk variants; (ii) identification and analysis of relationships between host genetic background and particular tumor mutations or types; and (iii) use of tumor molecular profiling data to reduce genetic heterogeneity or refine phenotypes for germline association studies. We also found descriptive studies that compared germline and tumor genomic variation in a gene or gene family, and papers describing research methods, data sources, or analytical tools. We identified a large set of tools and data resources that can be used to analyze and integrate data from both genomes. Finally, we discuss opportunities and challenges for cancer research that integrates germline and tumor genomics data.
Published by Oxford University Press 2014.
Figures
References
-
- Green E.D., et al. (2011). Charting a course for genomic medicine from base pairs to bedside. Nature, 470, 204–213 - PubMed
-
- Hindorff L., et al. (2013). A Catalog of Published Genome-Wide Association Studies. www.genome.gov/gwastudies (5 September 2013, date last accessed).
-
- (2013). The Cancer Genome Atlas. http://cancergenome.nih.gov/ (5 September 2013, date last accessed).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

