Learning a weighted meta-sample based parameter free sparse representation classification for microarray data
- PMID: 25115965
- PMCID: PMC4130588
- DOI: 10.1371/journal.pone.0104314
Learning a weighted meta-sample based parameter free sparse representation classification for microarray data
Abstract
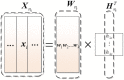
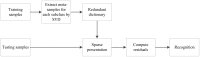
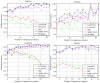
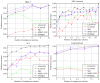
Sparse representation classification (SRC) is one of the most promising classification methods for supervised learning. This method can effectively exploit discriminating information by introducing a [Symbol: see text]1 regularization terms to the data. With the desirable property of sparisty, SRC is robust to both noise and outliers. In this study, we propose a weighted meta-sample based non-parametric sparse representation classification method for the accurate identification of tumor subtype. The proposed method includes three steps. First, we extract the weighted meta-samples for each sub class from raw data, and the rationality of the weighting strategy is proven mathematically. Second, sparse representation coefficients can be obtained by [Symnbol: see text]1 regularization of underdetermined linear equations. Thus, data dependent sparsity can be adaptively tuned. A simple characteristic function is eventually utilized to achieve classification. Asymptotic time complexity analysis is applied to our method. Compared with some state-of-the-art classifiers, the proposed method has lower time complexity and more flexibility. Experiments on eight samples of publicly available gene expression profile data show the effectiveness of the proposed method.
Conflict of interest statement
Figures



Similar articles
-
Maxdenominator Reweighted Sparse Representation for Tumor Classification.Sci Rep. 2017 Apr 10;7:46030. doi: 10.1038/srep46030. Sci Rep. 2017. PMID: 28393883 Free PMC article.
-
Metasample-based sparse representation for tumor classification.IEEE/ACM Trans Comput Biol Bioinform. 2011 Sep-Oct;8(5):1273-82. doi: 10.1109/TCBB.2011.20. IEEE/ACM Trans Comput Biol Bioinform. 2011. PMID: 21282864
-
Self-supervised sparse coding scheme for image classification based on low rank representation.PLoS One. 2018 Jun 20;13(6):e0199141. doi: 10.1371/journal.pone.0199141. eCollection 2018. PLoS One. 2018. PMID: 29924830 Free PMC article.
-
Sparse logistic regression with a L1/2 penalty for gene selection in cancer classification.BMC Bioinformatics. 2013 Jun 19;14:198. doi: 10.1186/1471-2105-14-198. BMC Bioinformatics. 2013. PMID: 23777239 Free PMC article.
-
Gene selection in cancer classification using sparse logistic regression with Bayesian regularization.Bioinformatics. 2006 Oct 1;22(19):2348-55. doi: 10.1093/bioinformatics/btl386. Epub 2006 Jul 14. Bioinformatics. 2006. PMID: 16844704
Cited by
-
Gene function prediction based on combining gene ontology hierarchy with multi-instance multi-label learning.RSC Adv. 2018 Aug 10;8(50):28503-28509. doi: 10.1039/c8ra05122d. eCollection 2018 Aug 7. RSC Adv. 2018. PMID: 35542493 Free PMC article.
-
Protein Sub-Nuclear Localization Based on Effective Fusion Representations and Dimension Reduction Algorithm LDA.Int J Mol Sci. 2015 Dec 19;16(12):30343-61. doi: 10.3390/ijms161226237. Int J Mol Sci. 2015. PMID: 26703574 Free PMC article.
-
Fisher Discrimination Regularized Robust Coding Based on a Local Center for Tumor Classification.Sci Rep. 2018 Jun 14;8(1):9152. doi: 10.1038/s41598-018-27364-7. Sci Rep. 2018. PMID: 29904059 Free PMC article.
-
Maxdenominator Reweighted Sparse Representation for Tumor Classification.Sci Rep. 2017 Apr 10;7:46030. doi: 10.1038/srep46030. Sci Rep. 2017. PMID: 28393883 Free PMC article.
-
Identification of DNA-protein Binding Sites through Multi-Scale Local Average Blocks on Sequence Information.Molecules. 2017 Nov 28;22(12):2079. doi: 10.3390/molecules22122079. Molecules. 2017. PMID: 29182548 Free PMC article.
References
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, et al. (2000) Distinct types of diffuse large b-cell lymphoma identified by gene expression profiling. Nature 403: 503–511. - PubMed
-
- West M (2003) Bayesian factor regression models in the large p, small n paradigm. Bayesian statistics 7: 723–732.
-
- Liu B, Fang B, Liu X, Chen J, Huang Z (2013) Large margin subspace learning for feature selection. Pattern Recognition.
-
- Cai D, He X, Zhou K, Han J, Bao H (2007) Locality sensitive discriminant analysis. In: IJCAI. pp. 708–713.
-
- Sugiyama M (2006) Local fisher discriminant analysis for supervised dimensionality reduction. In: Proceedings of the 23rd international conference on Machine learning ACM, pp. 905–912.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

